Final Presentation 11 16 2012 Sample Preparation Nextera





- Slides: 16

Final Presentation 11 -16 -2012

Sample Preparation • • Nextera Tru. Seq Ribo. Zero Strand Specific Clontech sm. RNA 16 s

Nextera • Quantification issues Nextera libraries consistently have lower q. PCR concentrations compared to BA; usually about half as concentrated • Illumina Sequences

My Guess?

Nextera Sequences • CTTAATGATACGGCGACCACCGAGATCTACACTAGATCGC TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGTCGTGA TGGACTGCCGTAACGGCGACCTGCTGTGCATGGCCTCCGC GCCGAGCTTCGACGCCAACCGGTTCGTGCGGGGGCTGTC CGGTCCTGAGTACAAGGCCCTGGCCGAGTACGAGCGCAA GCCGCTGCTCGACAAGTCGATGACCGGCCTGTTTCCGCCC GGCTCGACCTTCAAGCCCACGGTGGGTCTGGCCGCCCTG GCCGCCGGCATCGATCCCGAGGTCCGGGTCAACTGTCCG GGCAGCTGGTACTATGGCGGTCGGGTGTGGCGTTGCTGG GAGAAGGGCGGCCACGGCCTGTCTCTTATACACATCTCCG AGCCCACGAGACTAAGGCGAATCTCGTATGCCGTCTTCTG CTTG

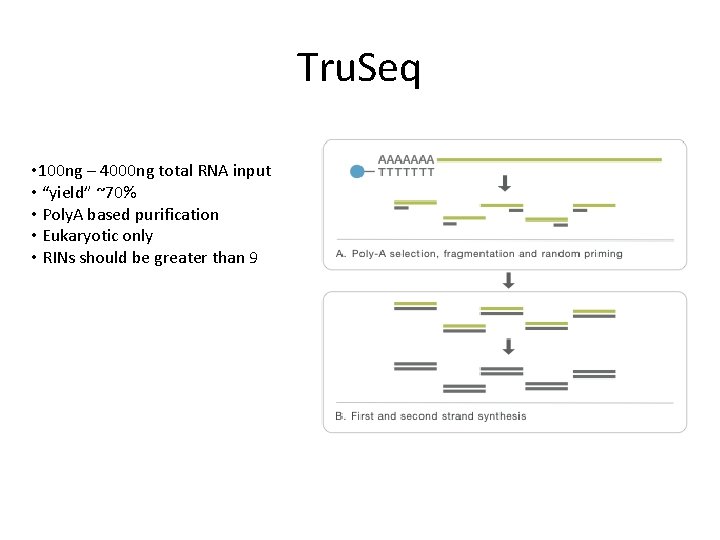
Tru. Seq • 100 ng – 4000 ng total RNA input • “yield” ~70% • Poly. A based purification • Eukaryotic only • RINs should be greater than 9

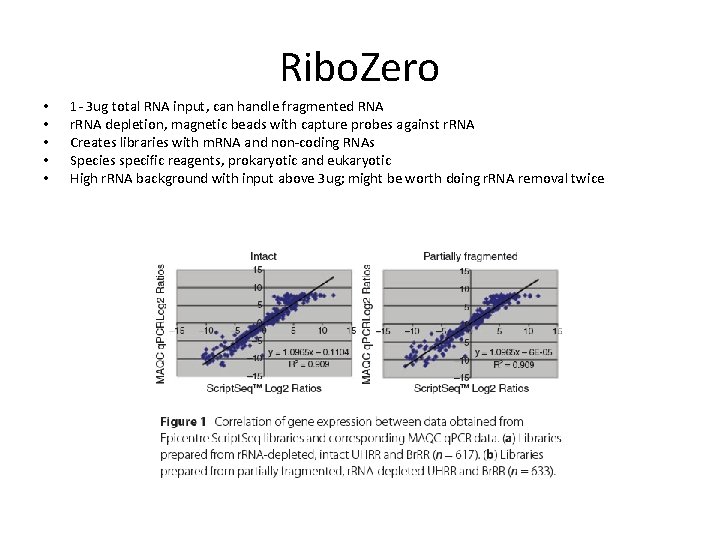
Ribo. Zero • • • 1 - 3 ug total RNA input, can handle fragmented RNA r. RNA depletion, magnetic beads with capture probes against r. RNA Creates libraries with m. RNA and non-coding RNAs Species specific reagents, prokaryotic and eukaryotic High r. RNA background with input above 3 ug; might be worth doing r. RNA removal twice

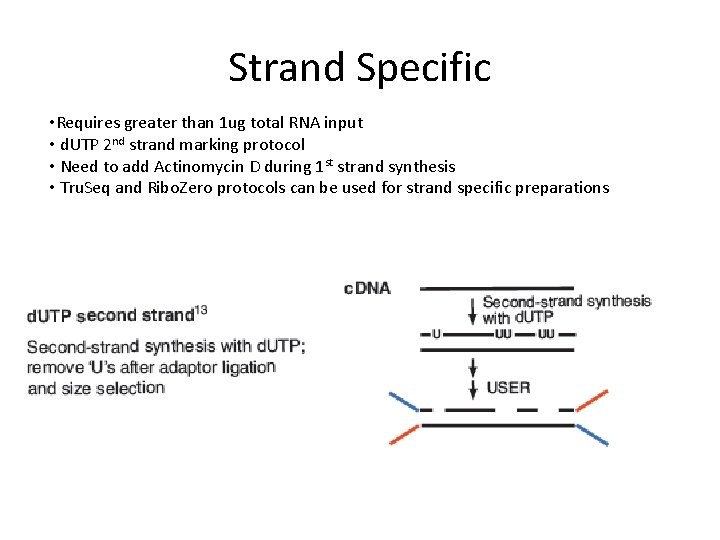
Strand Specific • Requires greater than 1 ug total RNA input • d. UTP 2 nd strand marking protocol • Need to add Actinomycin D during 1 st strand synthesis • Tru. Seq and Ribo. Zero protocols can be used for strand specific preparations

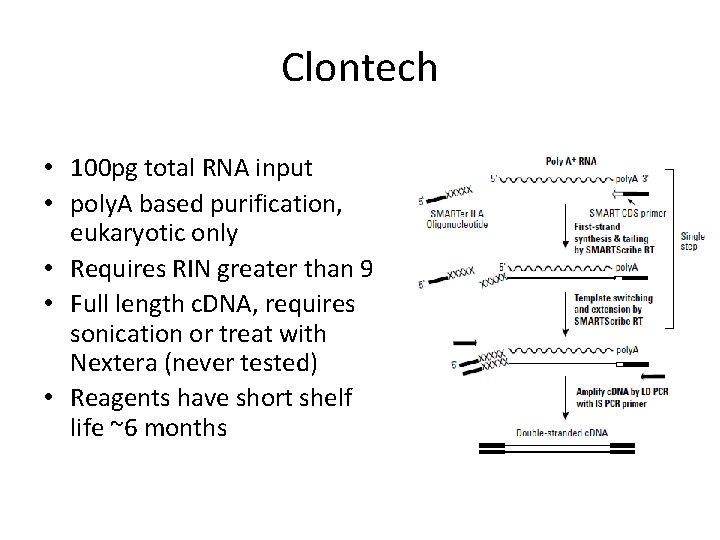
Clontech • 100 pg total RNA input • poly. A based purification, eukaryotic only • Requires RIN greater than 9 • Full length c. DNA, requires sonication or treat with Nextera (never tested) • Reagents have short shelf life ~6 months

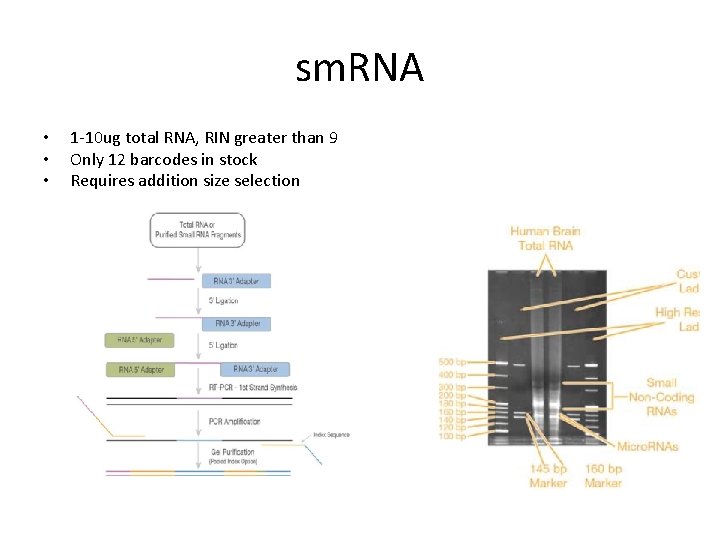
sm. RNA • • • 1 -10 ug total RNA, RIN greater than 9 Only 12 barcodes in stock Requires addition size selection

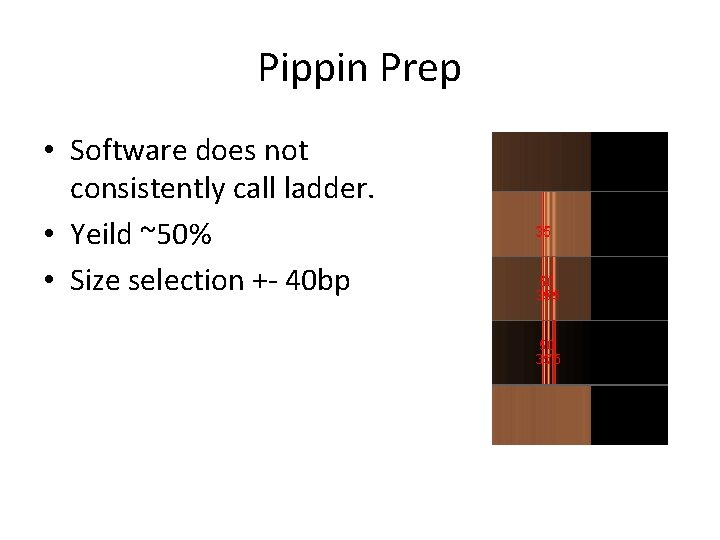
Pippin Prep • Software does not consistently call ladder. • Yeild ~50% • Size selection +- 40 bp

Metagenomics/16 s Sequencing • Metagenomics is an emerging field • Genomic analysis is applied to microbial communities rather than individual microbes • Bypasses the need to isolate and culture individual microbial species • Many microbes are resistant to culturing • Has potential medical uses.

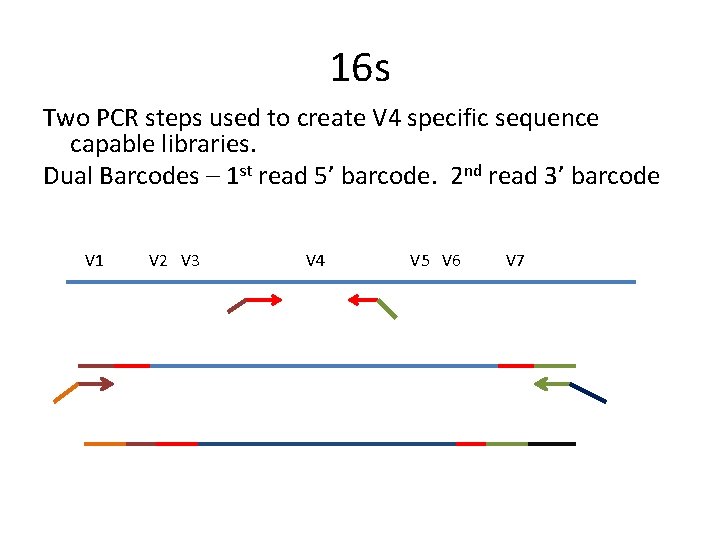
16 s Two PCR steps used to create V 4 specific sequence capable libraries. Dual Barcodes – 1 st read 5’ barcode. 2 nd read 3’ barcode V 1 V 2 V 3 V 4 V 5 V 6 V 7

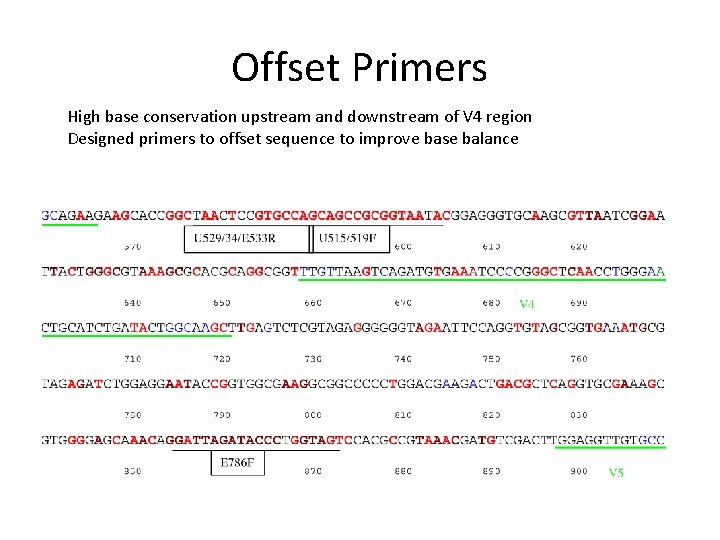
Offset Primers High base conservation upstream and downstream of V 4 region Designed primers to offset sequence to improve base balance

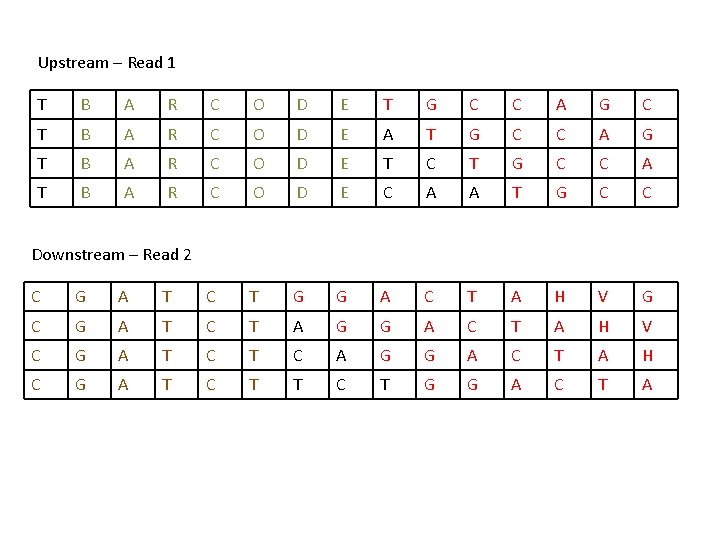
Upstream – Read 1 T B A R C O D E T G C C A G C T B A R C O D E A T G C C A G T B A R C O D E T C T G C C A T B A R C O D E C A A T G C C Downstream – Read 2 C G A T C T G G A C T A H V G C G A T C T A G G A C T A H V C G A T C A G G A C T A H C G A T C T G G A C T A

To Do • Optimize Ribo. Zero to reduce r. RNA contamination • Test ability to use Nextera instead of covartis fragmentation for clontech samples • Run 16 s offset samples to check base balance