Compiling Information and Inferring Useful Knowledge for Systems





- Slides: 14

Compiling Information and Inferring Useful Knowledge for Systems Biology by Text Mining the Literature Anália Lourenço IBB – Institute for Biotechnology and Bioengineering Centre of Biological Engineering, Bio. PSE group Universidade do Minho

Systems Biology does not investigate individual cellular components at a time, but the behaviour and relationships of all of the elements in a particular biological system while it is functioning. http: //www. personal. psu. edu/suk 211/blogs/susha nts_blogsphere/2009/07/system-biology-forpersonalized-medicine. html Bio. PSEg

Biomedical Literature Mining • The idea is to train computers to retrieve, read and interpret the text under processing: – what to read; – how to read; – what to do with the processed text. • Roughly speaking, we want to emulate human reading behaviour as closest as possible. – Learning domain-specific behaviour. – Aiming at delivering intuitive, comprehensible domain -specific knowledge. Bio. PSEg

Biomedical Literature Mining • Bio-entity tagging (mainly genes and proteins) • Gene –disease association • Enzyme-related information • Protein relations (binary relations • Pharmokinetics and interactions) • Metagenomics • Function annotation and • Automatic Information Retrieval: Retrieval • . . . localization relations • bulk access to Pub. Med contents; • Protein sequence (mutations, • retrieval of full-text documents; documents polymorphisms, modifications) • … • Acronym, synonym and term • Automatic Information Extraction: Extraction collection • document classification and clustering; clustering • extraction of biologically relevant • . . . information; information • knowledge inference • . . . Bio. PSEg

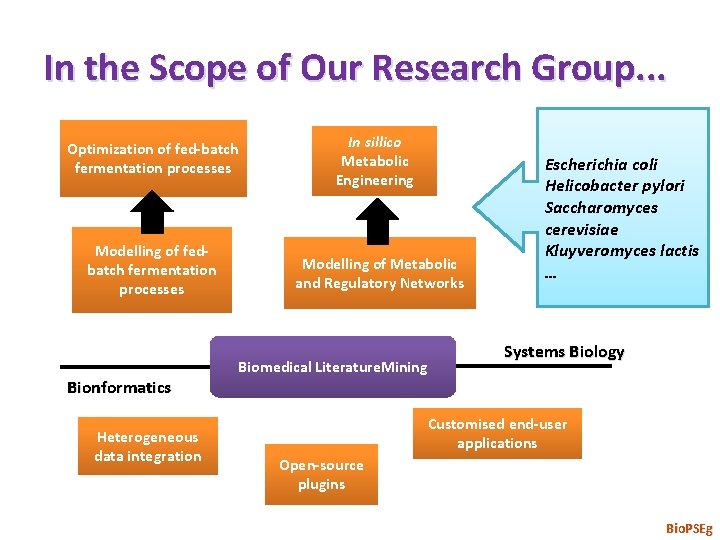
In the Scope of Our Research Group. . . Optimization of fed-batch fermentation processes In sillico Metabolic Engineering Modelling of fedbatch fermentation processes Modelling of Metabolic and Regulatory Networks Bionformatics Heterogeneous data integration Biomedical Literature. Mining Escherichia coli Helicobacter pylori Saccharomyces cerevisiae Kluyveromyces lactis … Systems Biology Customised end-user applications Open-source plugins Bio. PSEg

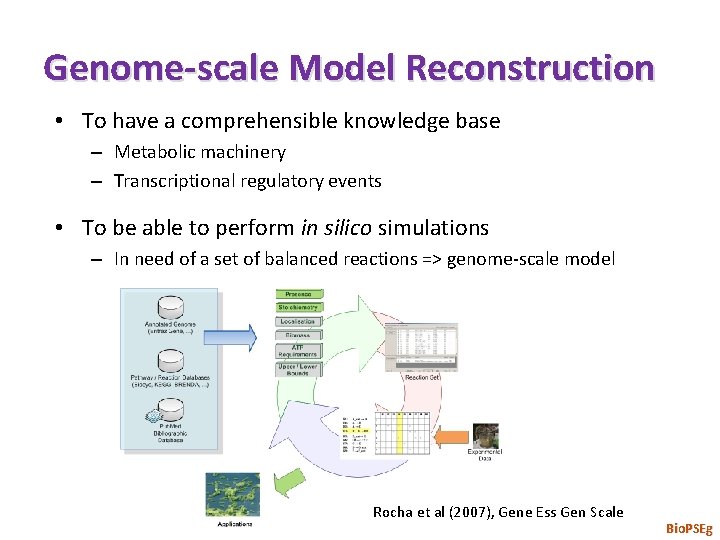
Genome-scale Model Reconstruction • To have a comprehensible knowledge base – Metabolic machinery – Transcriptional regulatory events • To be able to perform in silico simulations – In need of a set of balanced reactions => genome-scale model Rocha et al (2007), Gene Ess Gen Scale Bio. PSEg

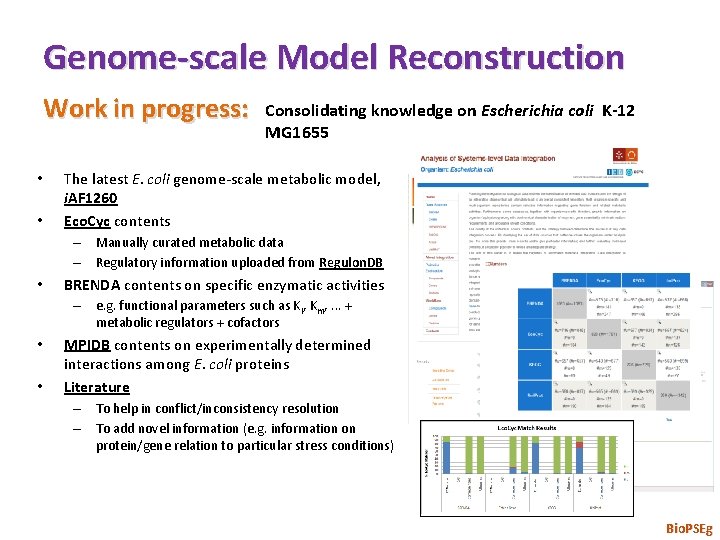
Genome-scale Model Reconstruction Work in progress: • • Consolidating knowledge on Escherichia coli K-12 MG 1655 The latest E. coli genome-scale metabolic model, i. AF 1260 Eco. Cyc contents – Manually curated metabolic data – Regulatory information uploaded from Regulon. DB • BRENDA contents on specific enzymatic activities – e. g. functional parameters such as Ki, Km, . . . + metabolic regulators + cofactors • • MPIDB contents on experimentally determined interactions among E. coli proteins Literature – To help in conflict/inconsistency resolution – To add novel information (e. g. information on protein/gene relation to particular stress conditions) Bio. PSEg

Genome-scale Model Reconstruction Work in progress: Reconstructing models for another organisms. . . • Helicobacter pylori • Kluyveromyces lactis • Streptococcus faecalis Bio. PSEg

Biomedical Literature Mining Work in progress: Mining the bibliome for a systematic review on the stringent response of the bacterium Escherichia coli Bio. PSEg

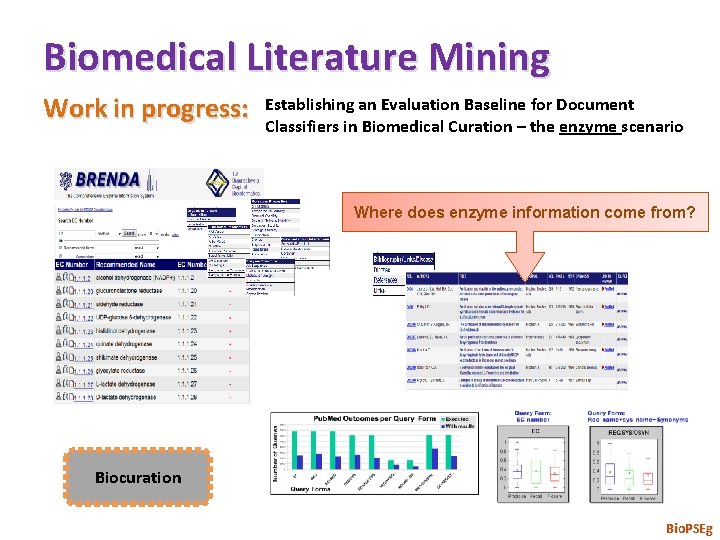
Biomedical Literature Mining Work in progress: Establishing an Evaluation Baseline for Document Classifiers in Biomedical Curation – the enzyme scenario Where does enzyme information come from? Biocuration Bio. PSEg

Biomedical Literature Mining Work in progress: Developing component modules for text mining services for biocuration Lourenço et al. (2010), Expert Systems With Applications , 37(4), 3444– 3453. Lourenço et al. (2009), J Biomed Inform. 42(4): 710 -20. Bio. PSEg

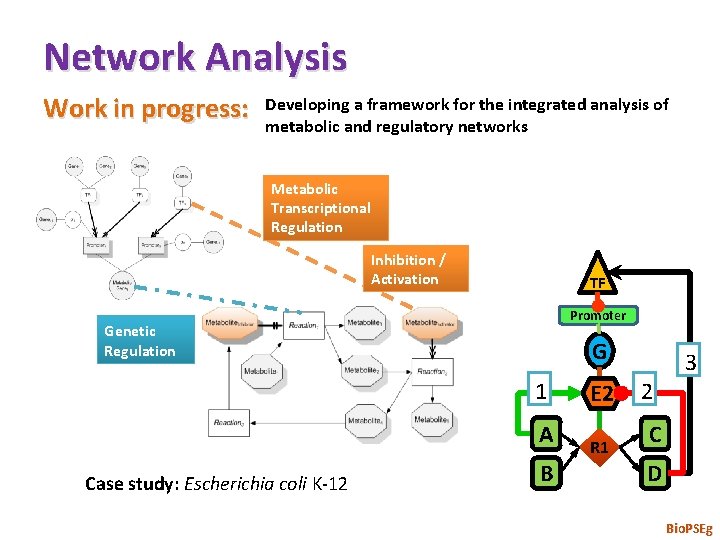
Network Analysis Work in progress: Developing a framework for the integrated analysis of metabolic and regulatory networks Metabolic Transcriptional Regulation Inhibition / Activation TF Promoter Genetic Regulation G 1 A Case study: Escherichia coli K-12 B E 2 R 1 2 3 C D Bio. PSEg

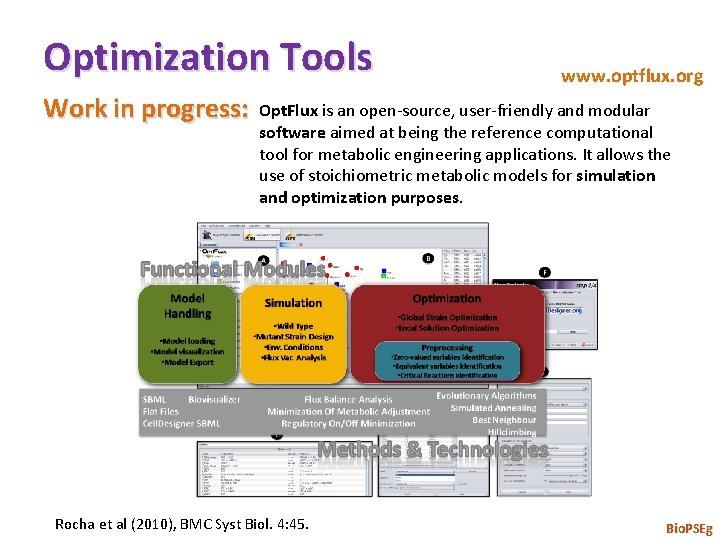
Optimization Tools Work in progress: www. optflux. org Opt. Flux is an open-source, user-friendly and modular software aimed at being the reference computational tool for metabolic engineering applications. It allows the use of stoichiometric metabolic models for simulation and optimization purposes. Rocha et al (2010), BMC Syst Biol. 4: 45. Bio. PSEg

HTTP: //BIOPSEG. DEB. UMINHO. PT/