Annotation and Visualization Doreen Ware Project Challenges Rapidly




- Slides: 20

Annotation and Visualization Doreen Ware

Project Challenges • Rapidly growing sequence data • Full annotation of all clones • New high-performance computing cluster • 2, 000 nodes • Scheduling system (Sun. Grid Engine) • NFS issues • Ens. EMBL Code Integration

• • Milestones www. maizesequence. org released Customized entry points of the Ensembl browser for the maize community. Adapted modules to the new compute cluster Blue Helix and automated gene predictions, MDR analysis, repeat masker Alignments of cereal sequence using Gramene Biopipe (needs to be automated) Transitioned from annotating Finished BACs to all BACs as they become available Blast Server FTP site DAS server (displaying Twinscan annotations)

Index Page

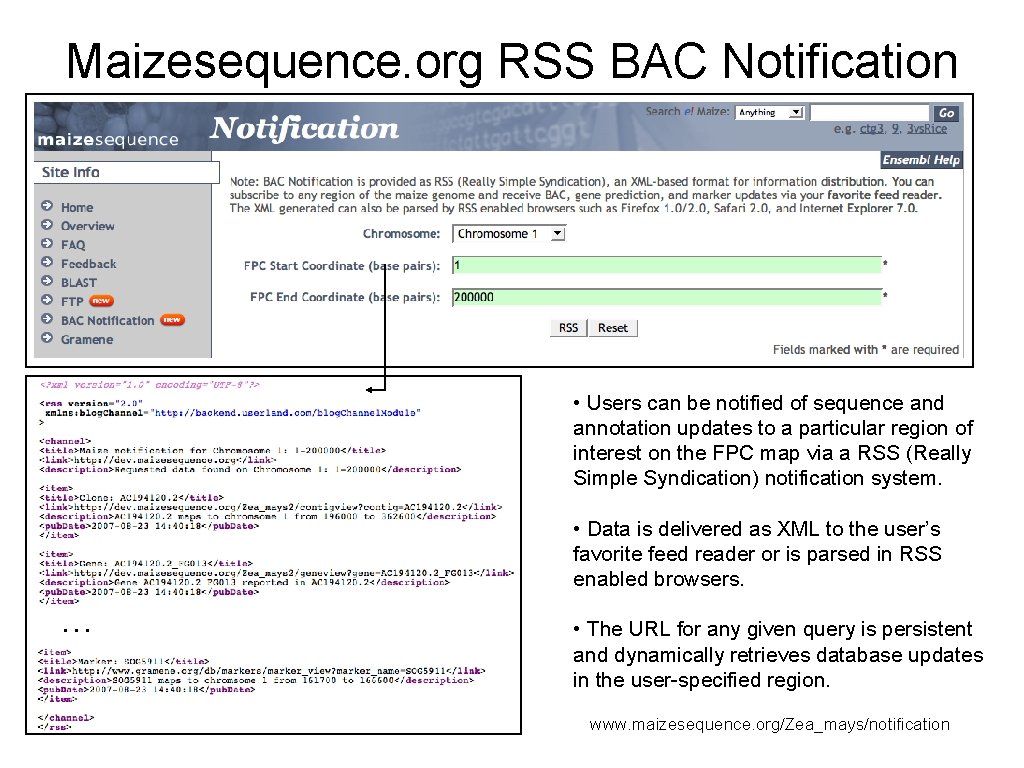
Maizesequence. org RSS BAC Notification • Users can be notified of sequence and annotation updates to a particular region of interest on the FPC map via a RSS (Really Simple Syndication) notification system. • Data is delivered as XML to the user’s favorite feed reader or is parsed in RSS enabled browsers. … • The URL for any given query is persistent and dynamically retrieves database updates in the user-specified region. www. maizesequence. org/Zea_mays/notification

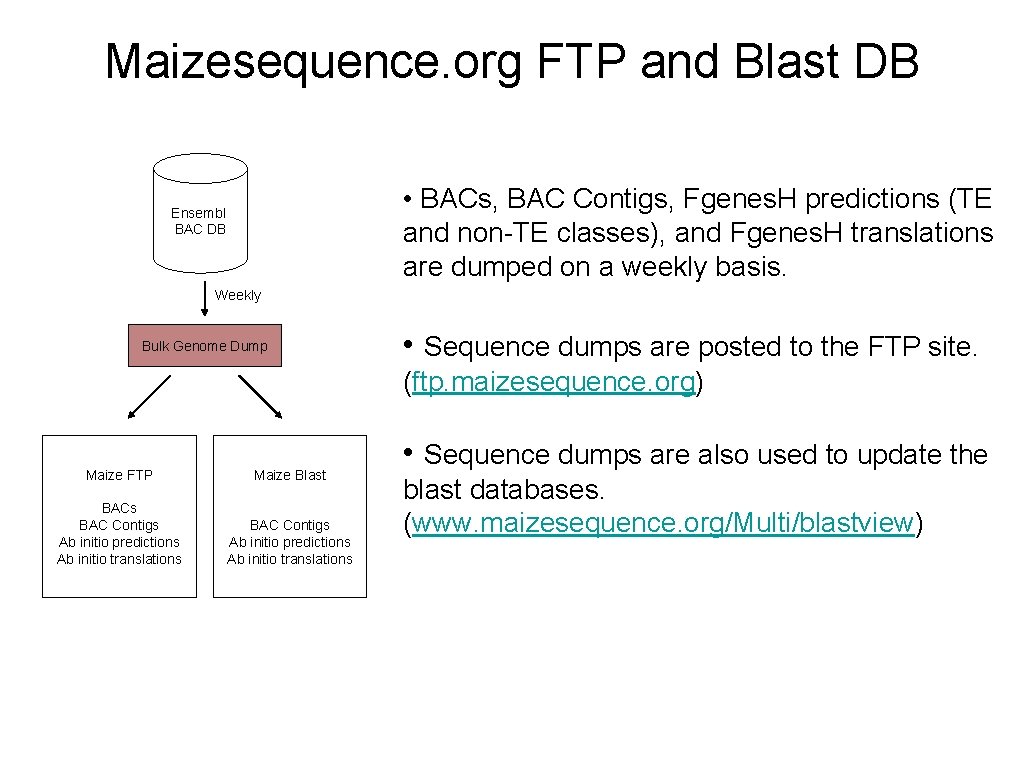
Maizesequence. org FTP and Blast DB • BACs, BAC Contigs, Fgenes. H predictions (TE and non-TE classes), and Fgenes. H translations are dumped on a weekly basis. Ensembl BAC DB Weekly Bulk Genome Dump • Sequence dumps are posted to the FTP site. (ftp. maizesequence. org) Maize FTP Maize Blast BACs BAC Contigs Ab initio predictions Ab initio translations • Sequence dumps are also used to update the blast databases. (www. maizesequence. org/Multi/blastview)

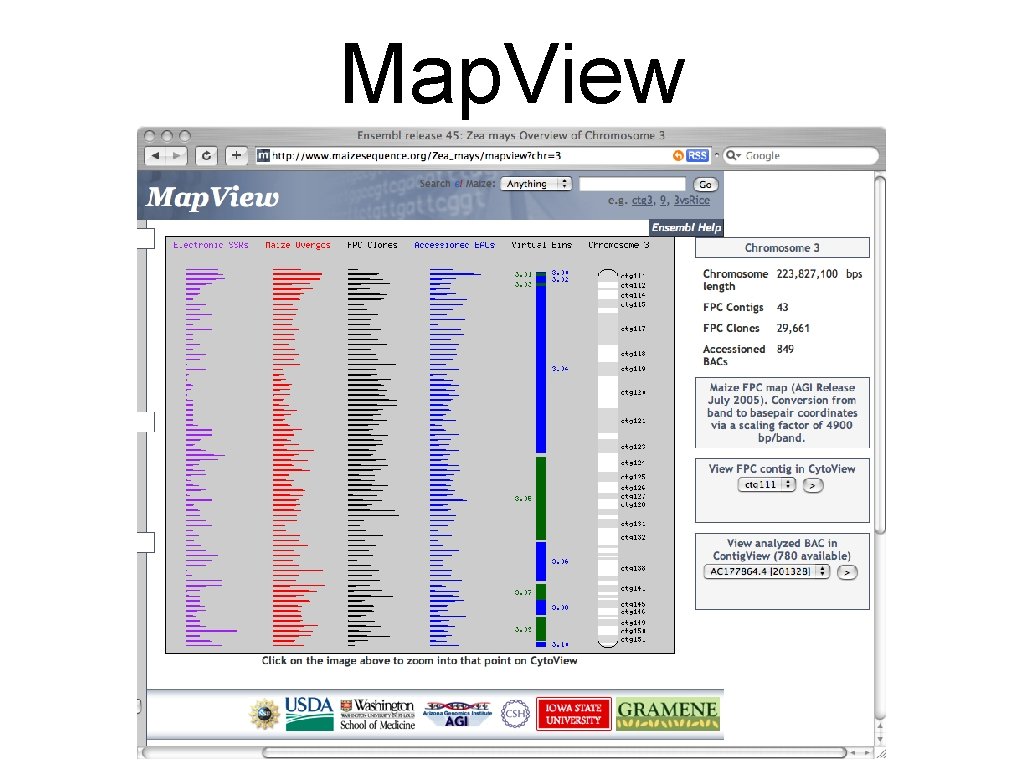
Map. View

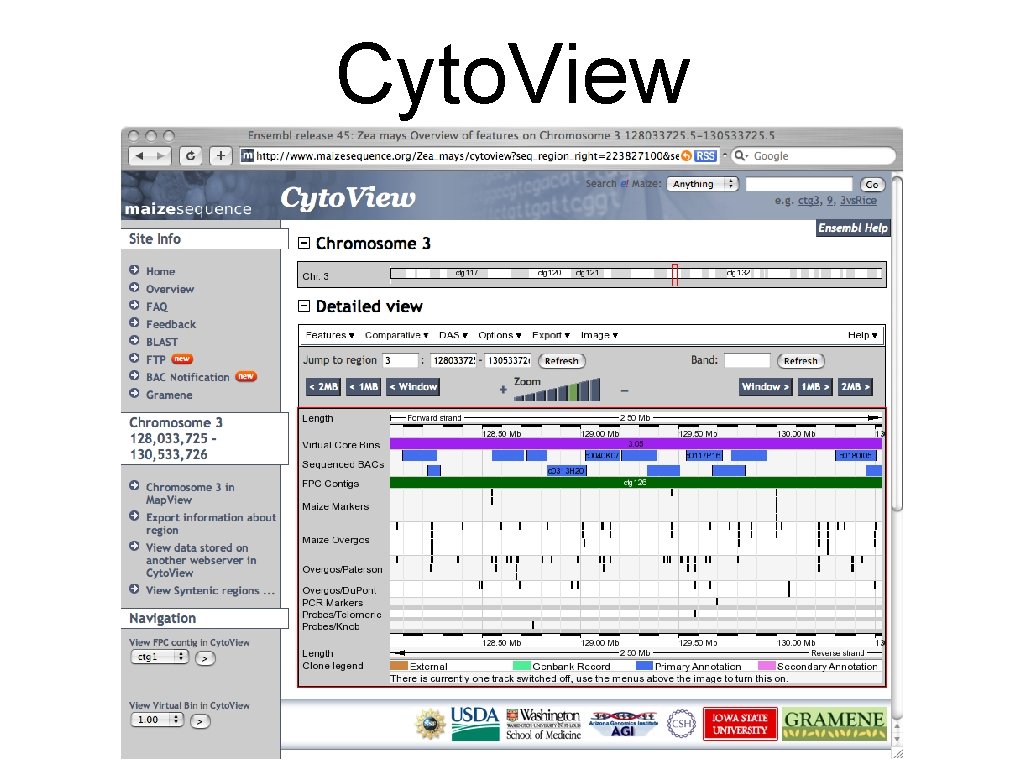
Cyto. View

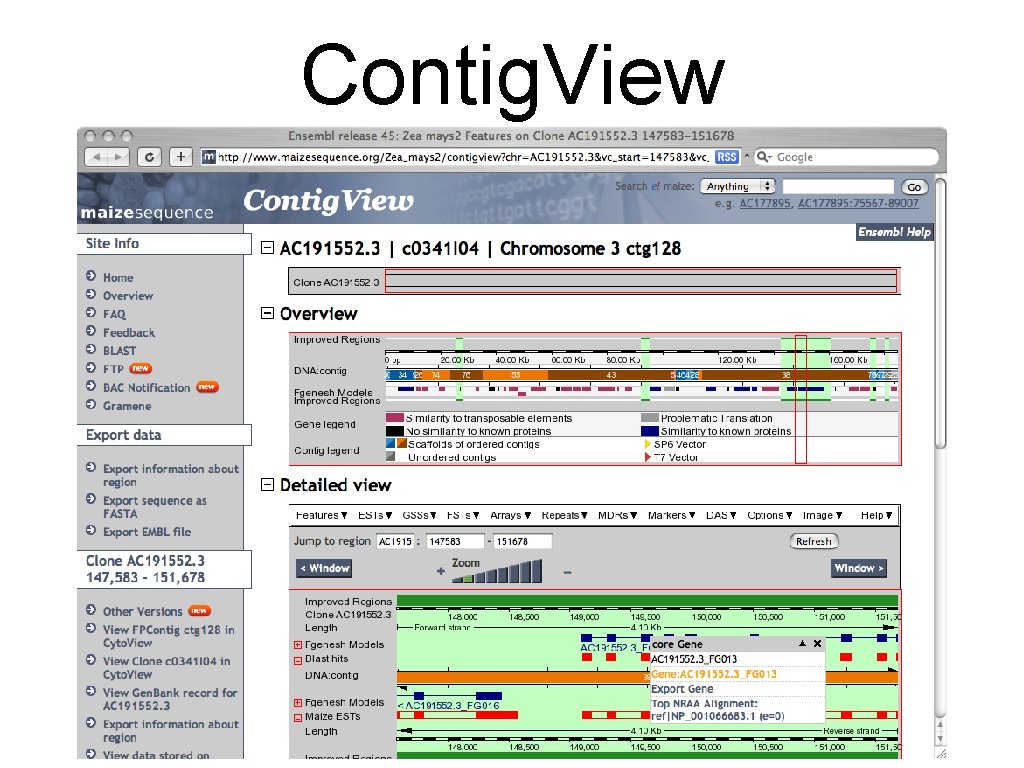
Contig. View

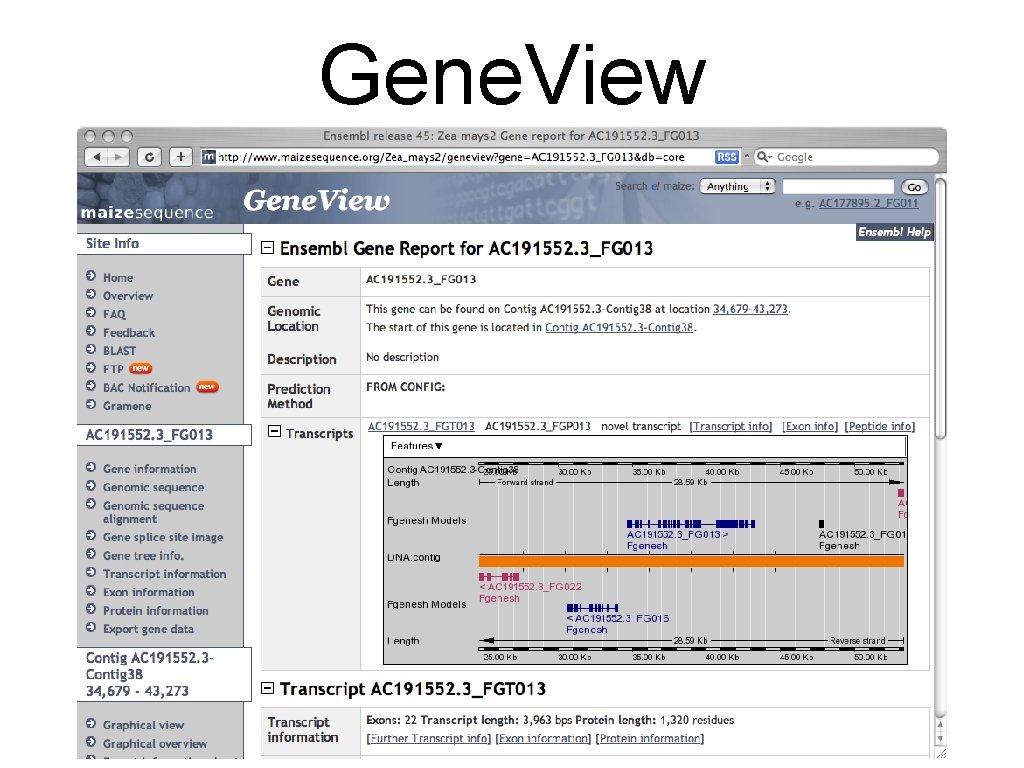
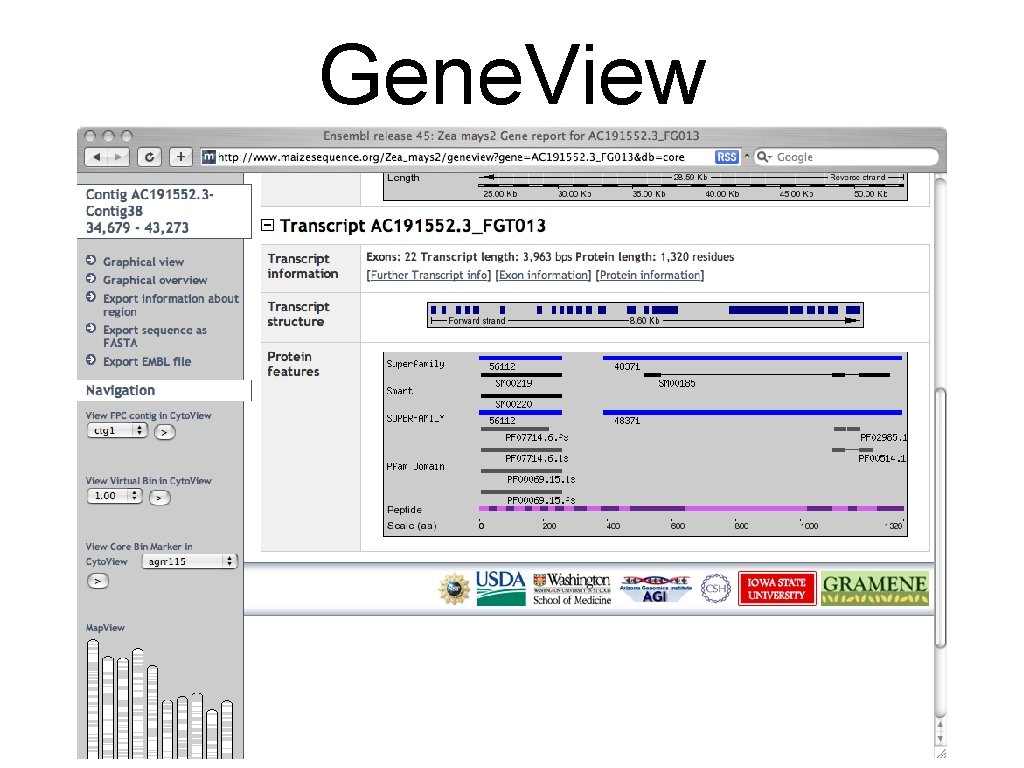
Gene. View

Gene. View

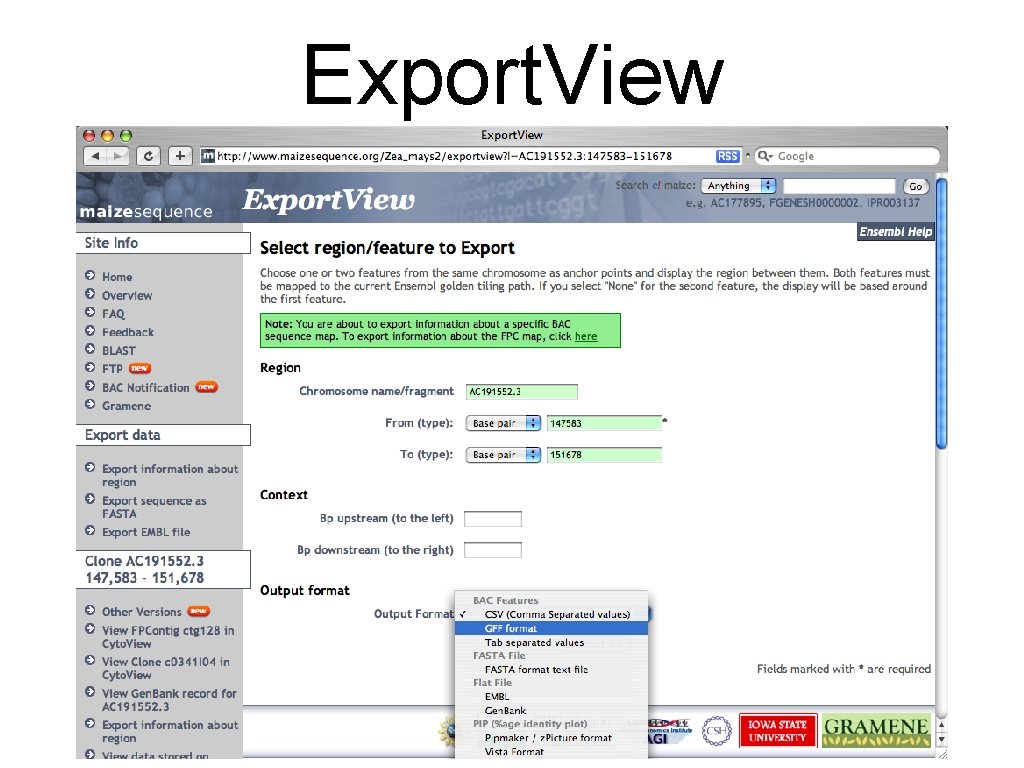
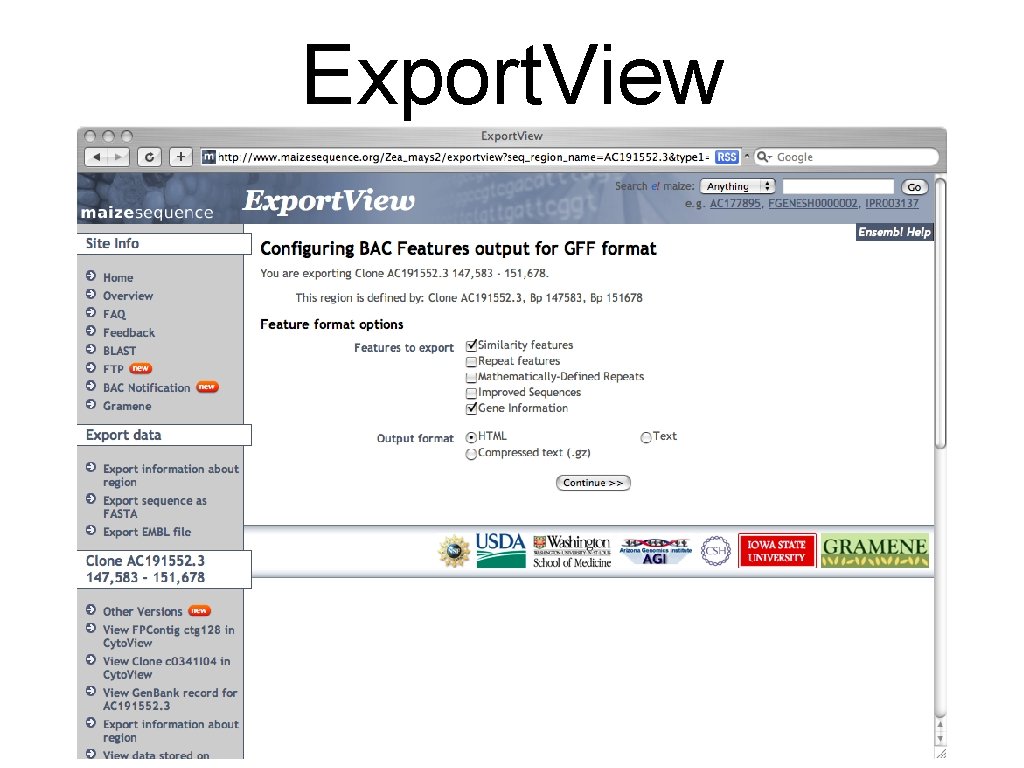
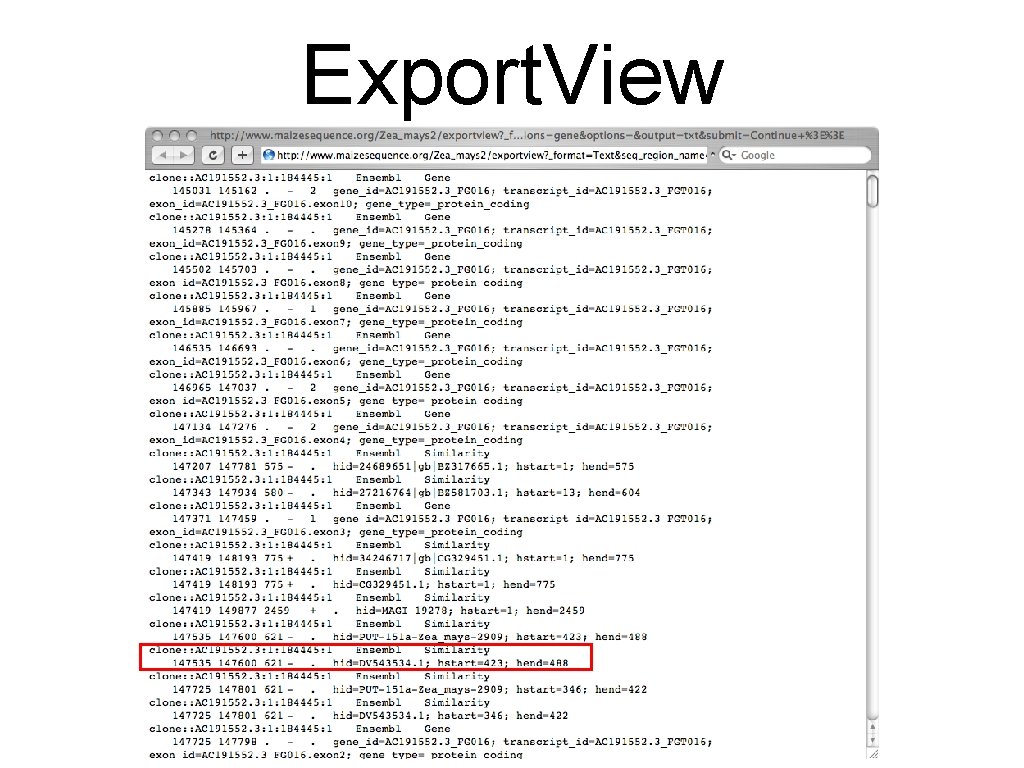
Export. View

Export. View

Export. View

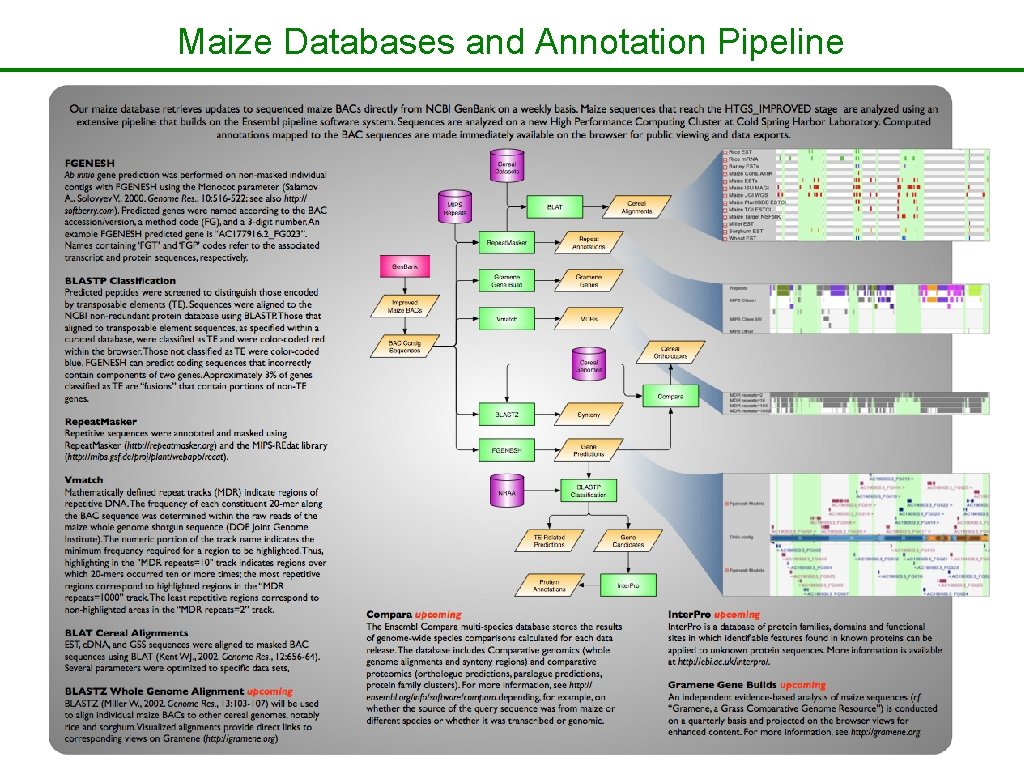
Maize Databases and Annotation Pipeline

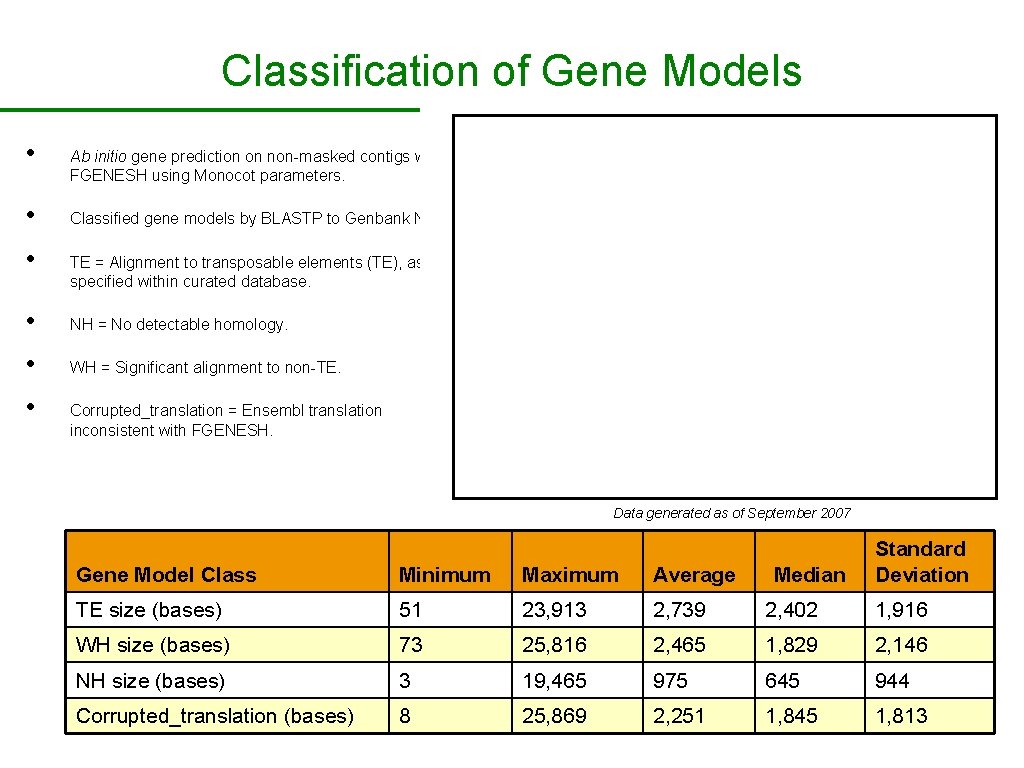
Classification of Gene Models • • • Ab initio gene prediction on non-masked contigs with FGENESH using Monocot parameters. Classified gene models by BLASTP to Genbank NRAA. TE = Alignment to transposable elements (TE), as specified within curated database. • NH = No detectable homology. • WH = Significant alignment to non-TE. • Corrupted_translation = Ensembl translation inconsistent with FGENESH. Data generated as of September 2007 Median Standard Deviation Gene Model Class Minimum Maximum Average TE size (bases) 51 23, 913 2, 739 2, 402 1, 916 WH size (bases) 73 25, 816 2, 465 1, 829 2, 146 NH size (bases) 3 19, 465 975 645 944 Corrupted_translation (bases) 8 25, 869 2, 251 1, 845 1, 813

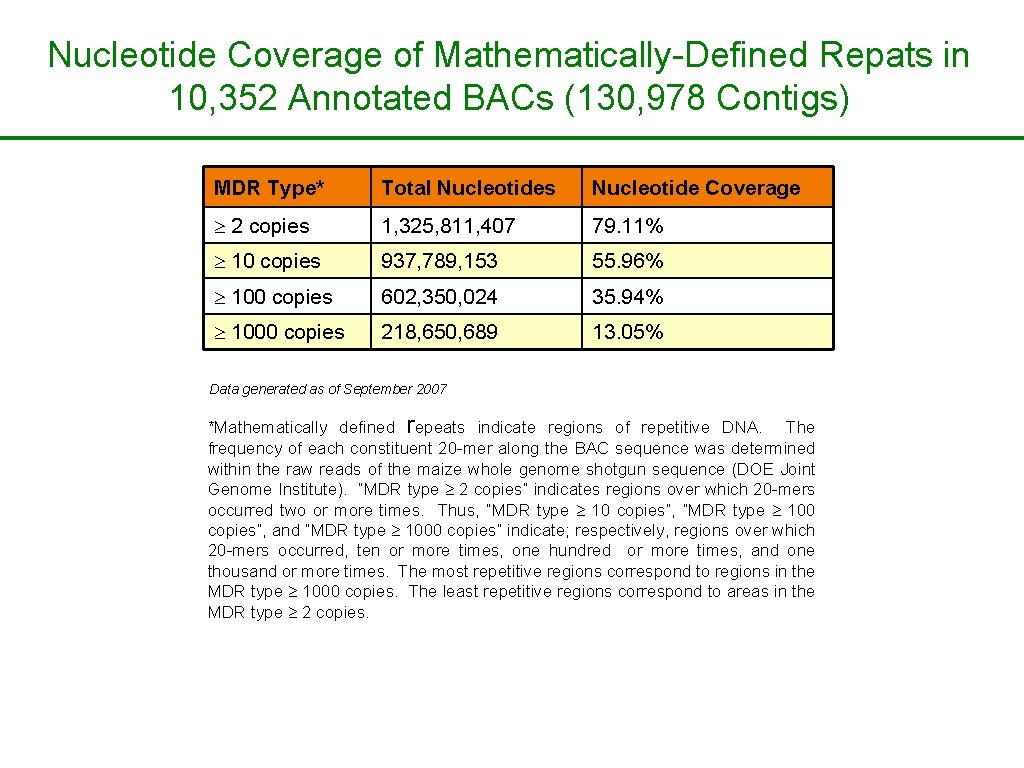
Nucleotide Coverage of Mathematically-Defined Repats in 10, 352 Annotated BACs (130, 978 Contigs) MDR Type* Total Nucleotides Nucleotide Coverage 2 copies 1, 325, 811, 407 79. 11% 10 copies 937, 789, 153 55. 96% 100 copies 602, 350, 024 35. 94% 1000 copies 218, 650, 689 13. 05% Data generated as of September 2007 *Mathematically defined repeats indicate regions of repetitive DNA. The frequency of each constituent 20 -mer along the BAC sequence was determined within the raw reads of the maize whole genome shotgun sequence (DOE Joint Genome Institute). “MDR type 2 copies” indicates regions over which 20 -mers occurred two or more times. Thus, “MDR type 10 copies”, “MDR type 100 copies”, and “MDR type 1000 copies” indicate; respectively, regions over which 20 -mers occurred, ten or more times, one hundred or more times, and one thousand or more times. The most repetitive regions correspond to regions in the MDR type 1000 copies. The least repetitive regions correspond to areas in the MDR type 2 copies.

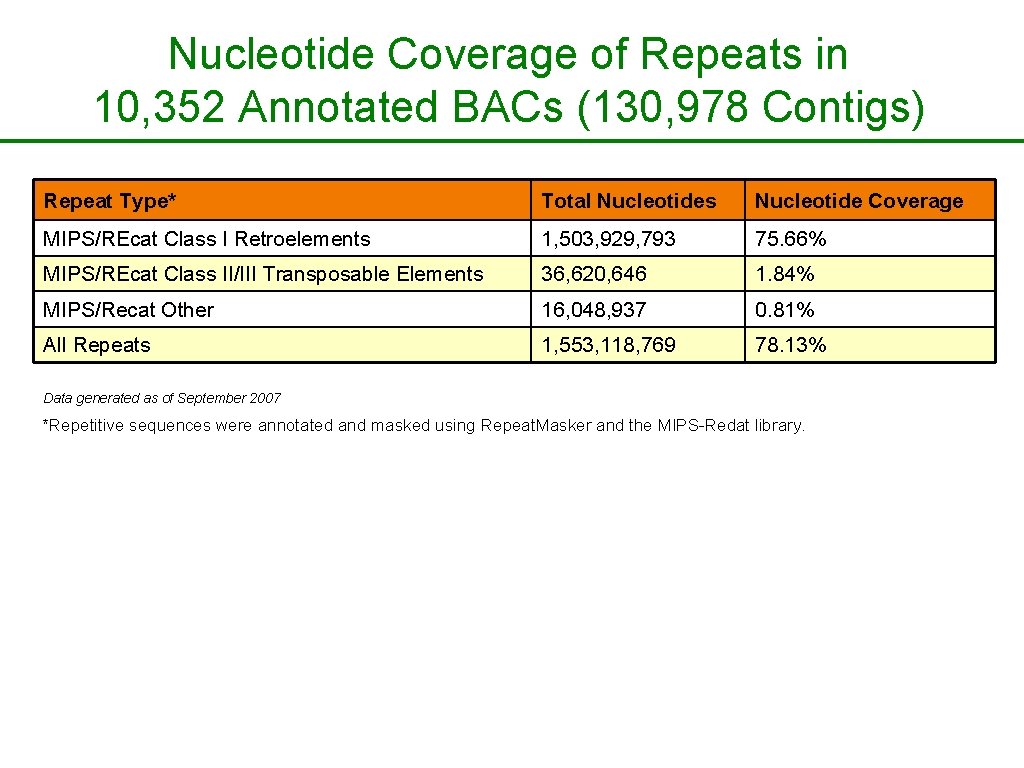
Nucleotide Coverage of Repeats in 10, 352 Annotated BACs (130, 978 Contigs) Repeat Type* Total Nucleotides Nucleotide Coverage MIPS/REcat Class I Retroelements 1, 503, 929, 793 75. 66% MIPS/REcat Class II/III Transposable Elements 36, 620, 646 1. 84% MIPS/Recat Other 16, 048, 937 0. 81% All Repeats 1, 553, 118, 769 78. 13% Data generated as of September 2007 *Repetitive sequences were annotated and masked using Repeat. Masker and the MIPS-Redat library.

Outreach and Collaborations • Maize. GDB • EBI Ens. EMBL • Gramene • Maize Array Working Group • Maize Optical Map • Transposon Annotation • TWINSCAN • Vmatch • Student Annotation (Howard Hughes)

Objectives for Year 3 • • • Whole Genome Alignments for rice, maize and arabidopsis Evidence based gene builds Gramene modified Ensembl pipeline and FGENESH++ in combiner mode Bio. Mart for complex query generation Whole Genome Alignments for rice, maize and arabidopsis Synteny. View based on whole genome alignment Transition from Gramene Biopipe -> Ensembl Exonerate pipeline to automate sequence alignments Annotation of non-coding RNA using t. RNAScan and micro. RNA Gene Ontology using dbxref pipeline Incorporation in Gramene Compara builds; Gene. Tree view My. SQL Database dumps Tutorials for website using Camptasia Submit paper on MDR analysis Shiran Pasternak, Apurva Narechania, Linda Mc. Mahan, Joshua Stein