2 Hydroxyethyl Triphenylphosphonium Chloride Large MoleculeSmall Molecule Interactions





- Slides: 14

(2 -Hydroxyethyl) Triphenylphosphonium Chloride Large Molecule/Small Molecule Interactions Rhode Island College July 14 -18, 2008 Bailey Sarber, Samantha Barrus Westerly High School

TPPEO • Molecular Formula-- HOCH 2 P(C 6 H 5)3 Cl • Molecular Weight— 342. 79 AMU • Physical State— white, crystalline powder • Solubility in Water– not available • LD 50– not available

Electrophoresis Well 2 - kb Ladder Well 3 - Plasmid DNA sample uncut (5μL) - 6300 base pairs Well 4 - Plasmid DNA sample uncut (5μL) with 1 µL of TPPEO- 6300 base pairs Well 5 - Plasmid DNA sample linearized (5μL) - 6200 base pairs Well 6 - Plasmid DNA sample linearized (5μL) with 1 µL of TPPEO- 6200 base pairs Possible Conclusions: • The doping agent failed to bind with the DNA samples. • The agent bound, but had no effect. • The agent’s concentration was too low. Final conclusion: Additional tests yielded the same results despite higher concentrations of TPPEO doping agent (3 µL and 5 µL).

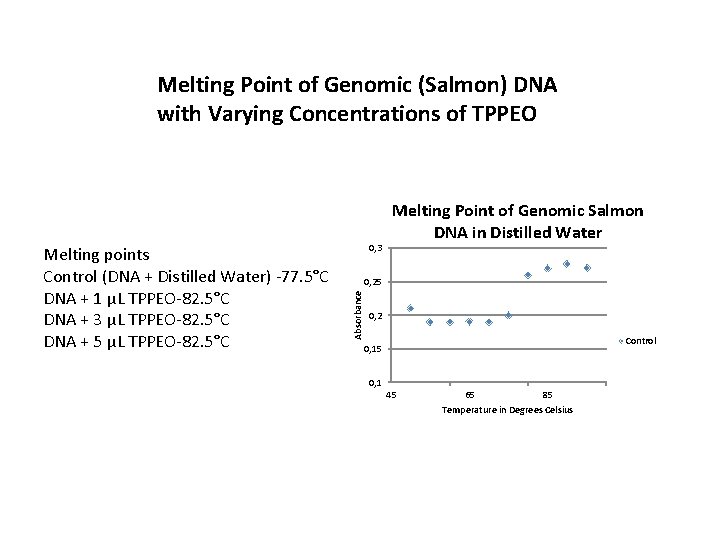
Melting Point of Genomic (Salmon) DNA with Varying Concentrations of TPPEO 0, 3 0, 25 Absorbance Melting points Control (DNA + Distilled Water) -77. 5°C DNA + 1 μL TPPEO-82. 5°C DNA + 3 μL TPPEO-82. 5°C DNA + 5 μL TPPEO-82. 5°C Melting Point of Genomic Salmon DNA in Distilled Water 0, 2 Control 0, 15 0, 1 45 65 85 Temperature in Degrees Celsius

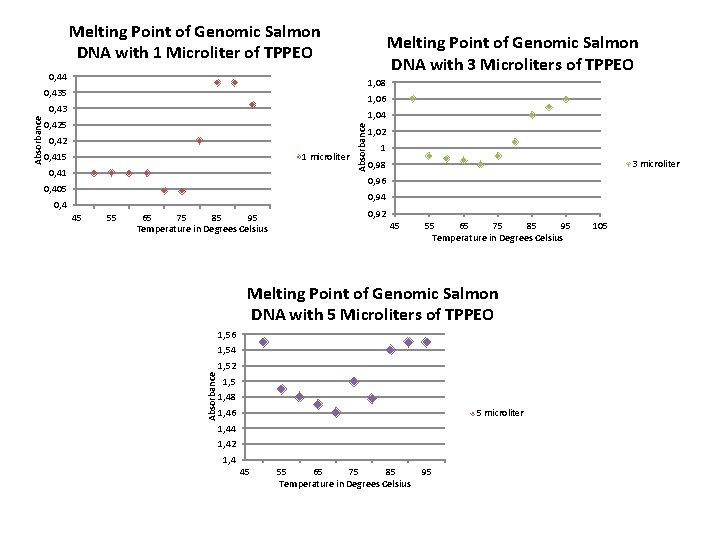
Melting Point of Genomic Salmon DNA with 1 Microliter of TPPEO Melting Point of Genomic Salmon DNA with 3 Microliters of TPPEO 0, 44 1, 08 0, 435 1, 06 1, 04 0, 425 0, 42 0, 415 1 microliter 0, 41 Absorbance 0, 43 1, 02 1 3 microliter 0, 98 0, 96 0, 405 0, 94 0, 4 55 65 75 85 95 Temperature in Degrees Celsius 0, 92 45 55 65 75 85 95 Temperature in Degrees Celsius Melting Point of Genomic Salmon DNA with 5 Microliters of TPPEO 1, 56 1, 54 1, 52 Absorbance 45 1, 48 1, 46 5 microliter 1, 44 1, 42 1, 4 45 55 65 75 85 Temperature in Degrees Celsius 95 105

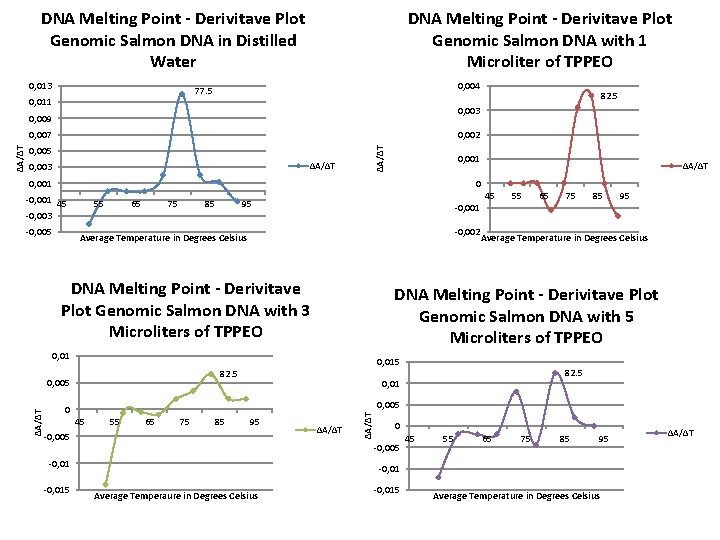
DNA Melting Point - Derivitave Plot Genomic Salmon DNA in Distilled Water 0, 013 DNA Melting Point - Derivitave Plot Genomic Salmon DNA with 1 Microliter of TPPEO 0, 004 77. 5 0, 011 0, 003 0, 009 0, 002 0, 005 ∆A/∆T 0, 003 ∆A/∆T 0, 007 ∆A/∆T 0, 001 ∆A/∆T 0 -0, 001 45 -0, 003 -0, 005 55 65 75 85 95 -0, 001 -0, 002 Average Temperature in Degrees Celsius DNA Melting Point - Derivitave Plot Genomic Salmon DNA with 3 Microliters of TPPEO 55 65 75 85 95 Average Temperature in Degrees Celsius 0, 015 82. 5 0, 005 82. 5 0, 01 45 55 65 75 85 95 -0, 005 ∆A/∆T 0, 005 0 0 -0, 005 -0, 015 45 DNA Melting Point - Derivitave Plot Genomic Salmon DNA with 5 Microliters of TPPEO 0, 01 ∆A/∆T 82. 5 45 55 65 75 85 95 -0, 01 Average Temperaure in Degrees Celsius -0, 015 Average Temperature in Degrees Celsius ∆A/∆T

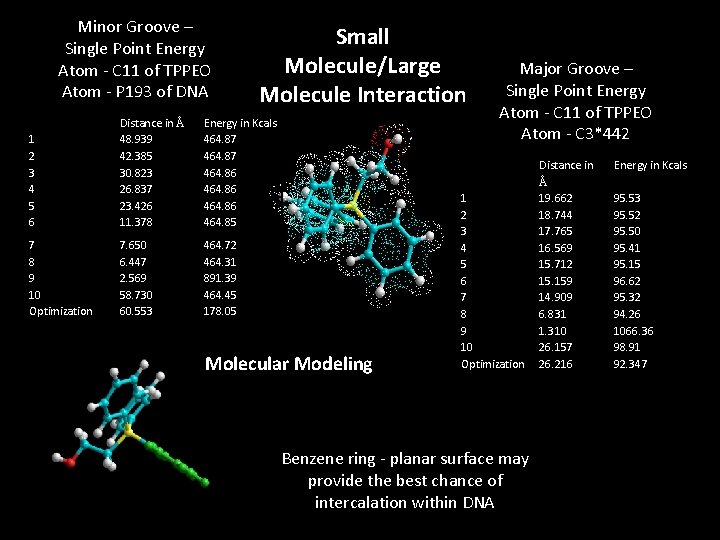
Minor Groove – Single Point Energy Atom - C 11 of TPPEO Atom - P 193 of DNA Small Molecule/Large Molecule Interaction 1 2 3 4 5 6 Distance in Å 48. 939 42. 385 30. 823 26. 837 23. 426 11. 378 Energy in Kcals 464. 87 464. 86 464. 85 7 8 9 10 Optimization 7. 650 6. 447 2. 569 58. 730 60. 553 464. 72 464. 31 891. 39 464. 45 178. 05 Molecular Modeling Major Groove – Single Point Energy Atom - C 11 of TPPEO Atom - C 3*442 1 2 3 4 5 6 7 8 9 10 Optimization Benzene ring - planar surface may provide the best chance of intercalation within DNA Distance in Å 19. 662 18. 744 17. 765 16. 569 15. 712 15. 159 14. 909 6. 831 1. 310 26. 157 26. 216 Energy in Kcals 95. 53 95. 52 95. 50 95. 41 95. 15 96. 62 95. 32 94. 26 1066. 36 98. 91 92. 347

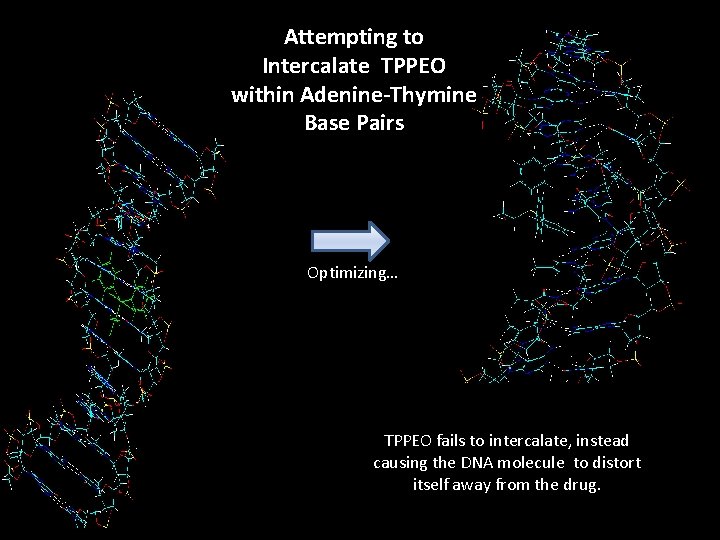
Attempting to Intercalate TPPEO within Adenine-Thymine Base Pairs Optimizing… TPPEO fails to intercalate, instead causing the DNA molecule to distort itself away from the drug.

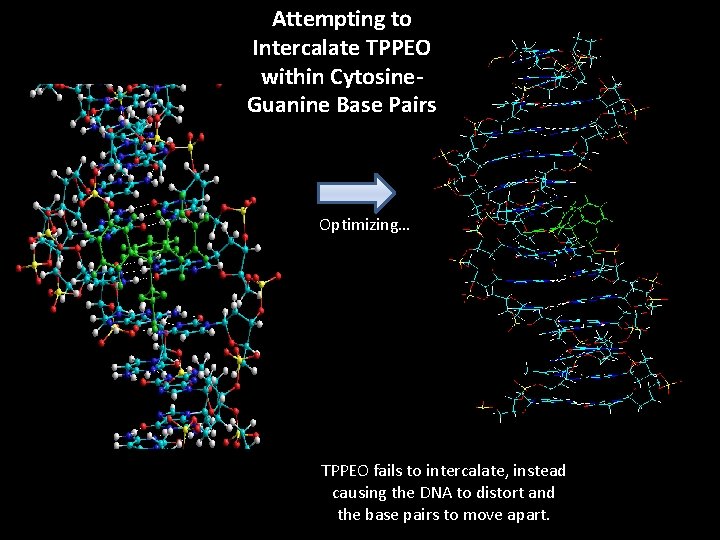
Attempting to Intercalate TPPEO within Cytosine. Guanine Base Pairs Optimizing… TPPEO fails to intercalate, instead causing the DNA to distort and the base pairs to move apart.

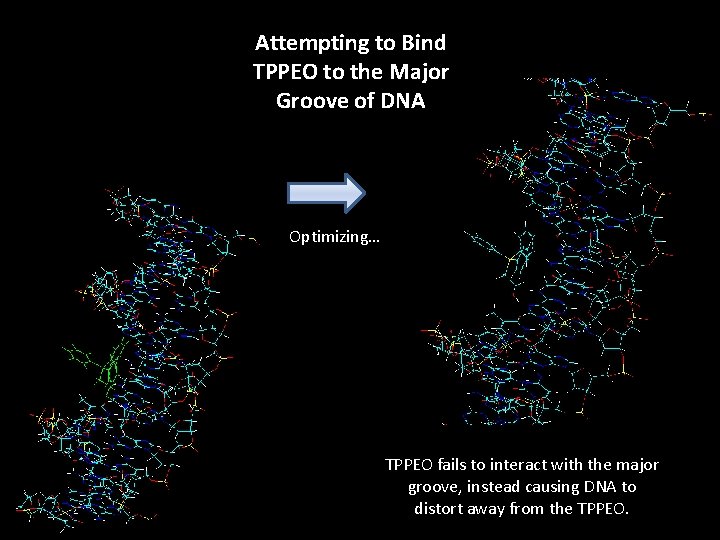
Attempting to Bind TPPEO to the Major Groove of DNA Optimizing… TPPEO fails to interact with the major groove, instead causing DNA to distort away from the TPPEO.

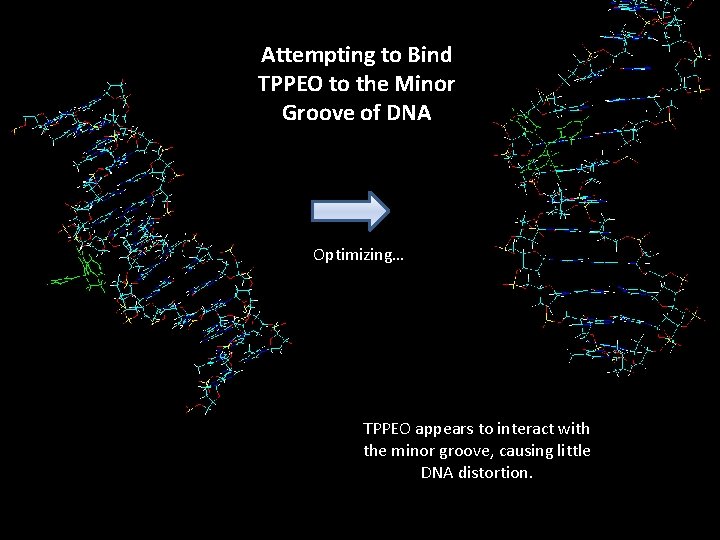
Attempting to Bind TPPEO to the Minor Groove of DNA Optimizing… TPPEO appears to interact with the minor groove, causing little DNA distortion.

Conclusions Electrophoresis- TPPEO did not affect the migration of the DNA through the gel. The drug either did not bind with DNA or it did bind with the DNA with no affect to the migration through the gel. Melting Point of Genomic DNA- There was a change in melting temperature between the control sample and those doped with various concentrations TPPEO including 1 μL, 3 μL and 5 μL. The possible shift was 5° Celsius indicating some sort of interaction between the drug and the DNA samples. Further testing is needed to substantiate an actual interaction. Molecular Modeling- Single point optimization between atoms on TPPEO and DNA revealed that when TPPEO approached the major groove on DNA the optimal energy level was 26. 216 Å. This suggests that the interaction based upon these two points is forced. Optimization on the minor groove had a greater overall distance than that of the major groove, at a distance of 60. 553Å. The energy level implies the two atoms do not interact well at the minor groove.

Adenine-Thymine Intercalation- TPPEO failed to intercalate within base pairs containing adenine-thymine sequences. The double hydrogen bonds connecting the two bases were not strong enough to withstand the distortion caused by the attempted intercalation. Cytosine-Guanine Intercalation-TPPEO again failed to intercalate between base pairs containing cytosine-guanine. These base pairs moved apart but not as far as the adenine-thyamine base pairs because three hydrogen bonds are stronger then two. Warning: Hyperchem has its limitations. Some sort of interaction appeared to take place in the minor groove of the DNA sequence when both molecules were optimized using the Amber program. This occurred despite the single point optimization not supporting the interaction. Single point optimization only configures two atoms but with so many configurations to test it would be realistically impossible to test them all using a program such as this. Intercalating within a DNA sequence has as many variables; base pair sequences, the size of the DNA molecule, and the location and configurations of the drug yield hypothetical results. Conclusion: Actual experimentation should be the primary source from which to draw conclusions. Recreating the experiment will quantify/support the data collected and verify the possible binding properties of TPPEO. Hyperchem is a tool, but not to be solely relied on.

Further Research… • Replicating the experiments • Confirming the assumed minor curve non-covalent bonding nature of TPPEO and DNA using X-Ray Crystallography • Investigate other molecules for possible DNA interaction ex: acetylsalicylic acid , ibuprofen, acetaminophen