ZO1 and Tight Junctions Cell Junctions http www







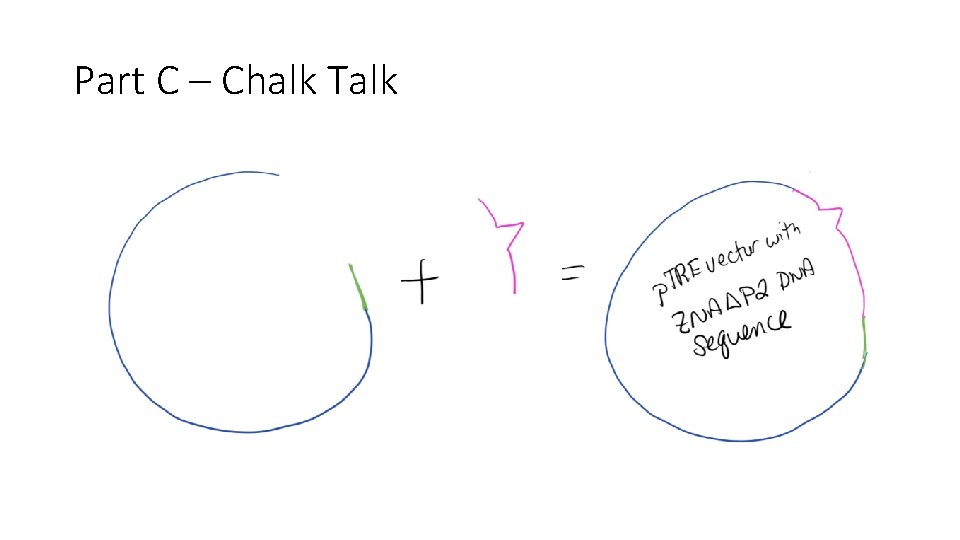
- Slides: 17

ZO-1 and Tight Junctions

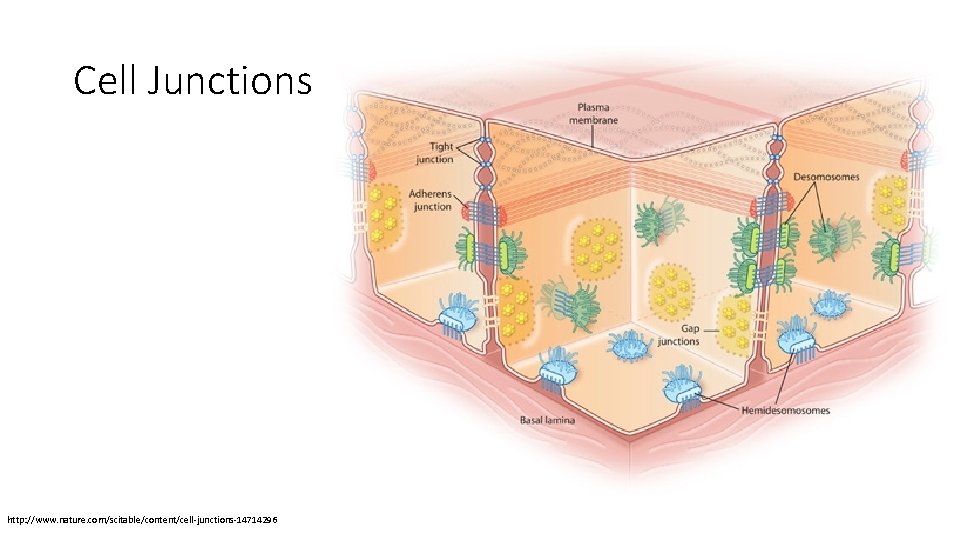
Cell Junctions http: //www. nature. com/scitable/content/cell-junctions-14714296

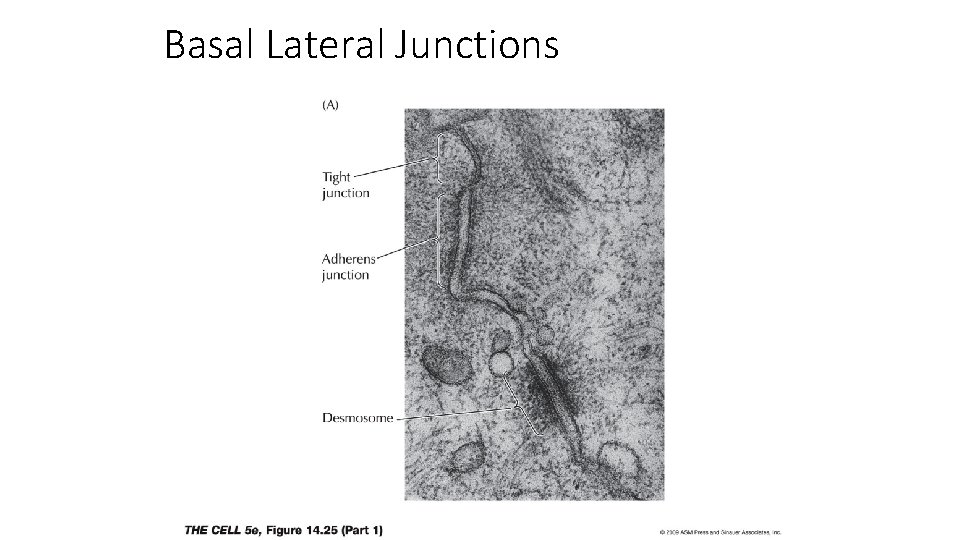
Basal Lateral Junctions

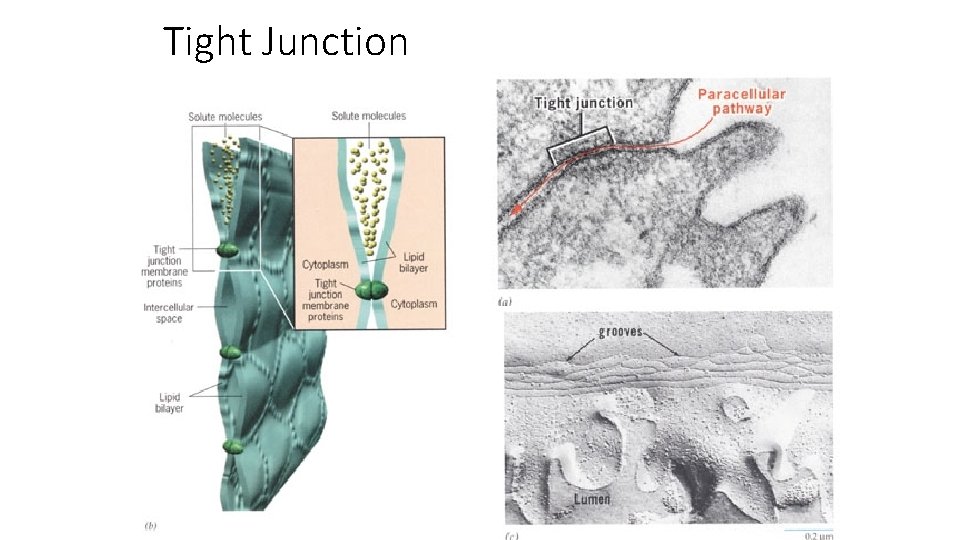
Tight Junction

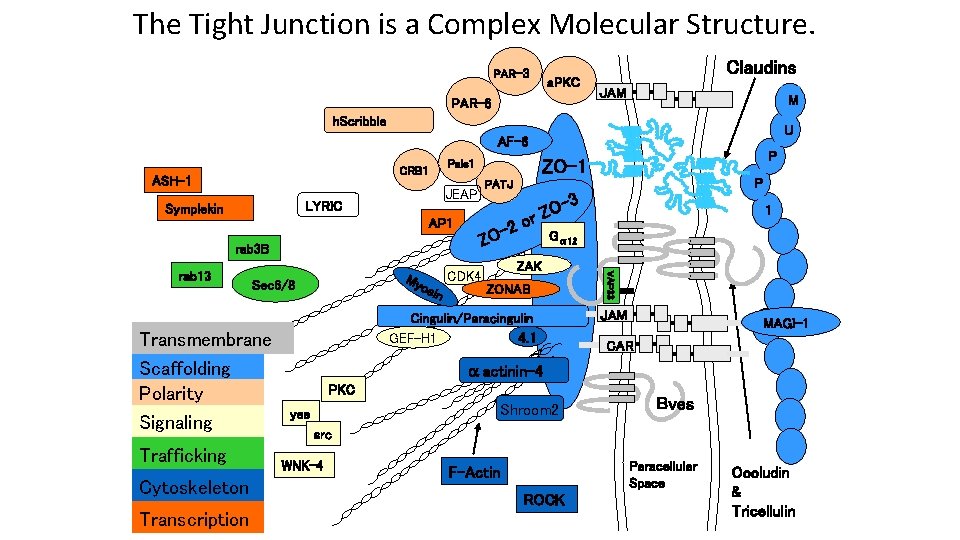
The Tight Junction is a Complex Molecular Structure. PAR-3 a. PKC PAR-6 Claudins JAM M h. Scribble U AF-6 ASH-1 JEAP LYRIC Symplekin AP 1 Sec 6/8 Transmembrane Scaffolding Polarity Signaling Trafficking Cytoskeleton Transcription My osi CDK 4 n -3 O Z or 1 Gα 12 ZAK ZONAB Cingulin/Paracingulin 4. 1 GEF-H 1 VAP 33 rab 13 P PATJ -2 ZO rab 3 B P ZO-1 Pals 1 CRB 1 JAM MAGI-1 CAR actinin-4 PKC Shroom 2 yes Bves src WNK-4 Paracellular Space F-Actin ROCK Occludin & Tricellulin

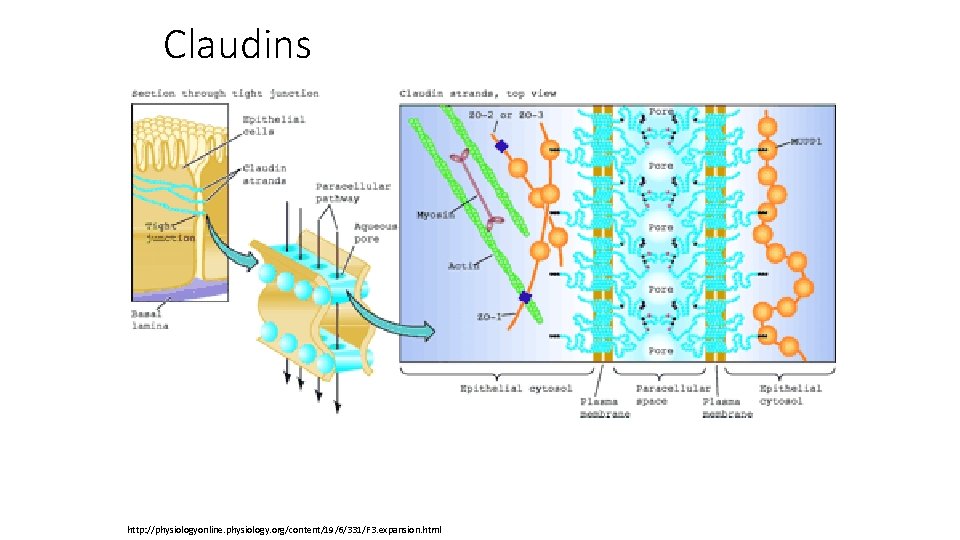
Claudins http: //physiologyonline. physiology. org/content/19/6/331/F 3. expansion. html

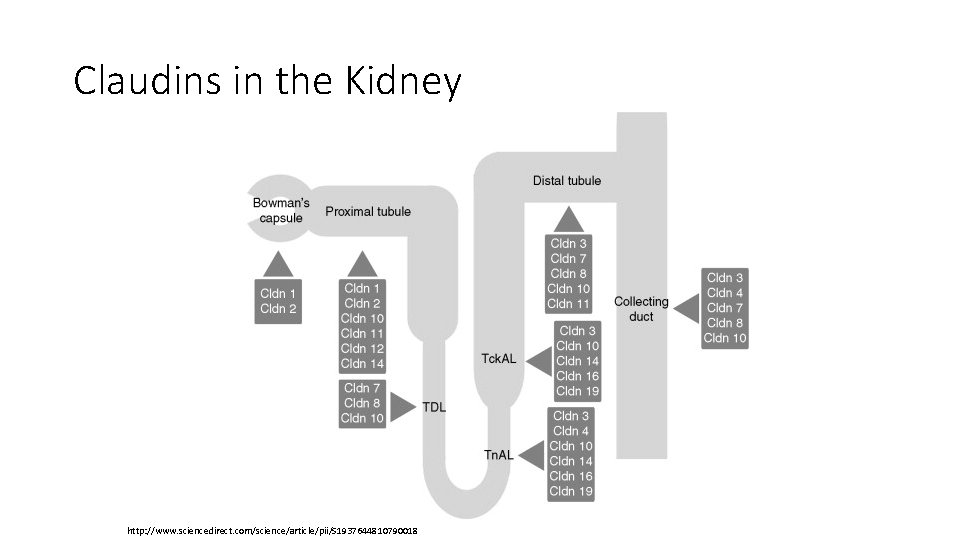
Claudins in the Kidney http: //www. sciencedirect. com/science/article/pii/S 1937644810790018

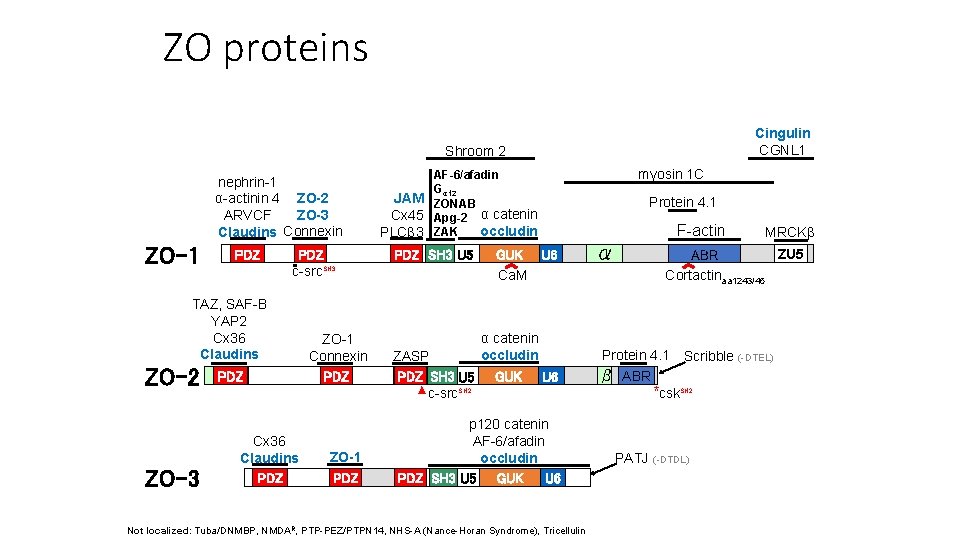
ZO proteins Cingulin CGNL 1 Shroom 2 nephrin-1 α-actinin 4 ZO-2 ZO-3 ARVCF Claudins Connexin ZO-1 PDZ c-src. SH 3 TAZ, SAF-B YAP 2 Cx 36 Claudins ZO-2 ZO-3 PDZ Cx 36 Claudins PDZ ZO-1 Connexin PDZ ZO-1 PDZ myosin 1 C AF-6/afadin Gα 12 JAM ZONAB Cx 45 Apg-2 α catenin occludin PLCβ 3 ZAK PDZ SH 3 U 5 ZASP PDZ SH 3 U 5 ▲ c-src. SH 2 Protein 4. 1 F-actin GUK U 6 Ca. M α catenin occludin GUK U 6 p 120 catenin AF-6/afadin occludin PDZ SH 3 U 5 GUK U 6 Not localized: Tuba/DNMBP, NMDAR, PTP-PEZ/PTPN 14, NHS-A (Nance-Horan Syndrome), Tricellulin α MRCKβ ZU 5 ABR Cortactinaa 1243/45 Protein 4. 1 Scribble (-DTEL) β ABR *csk. SH 2 PATJ (-DTDL)

Purpose of this study - to better understand the function of each ZO-1 binding domain Purpose of this lab - to walk students through the process necessary to do a protein structure/function project - to demonstrate that science can be “messy” and results are not always cut and dry

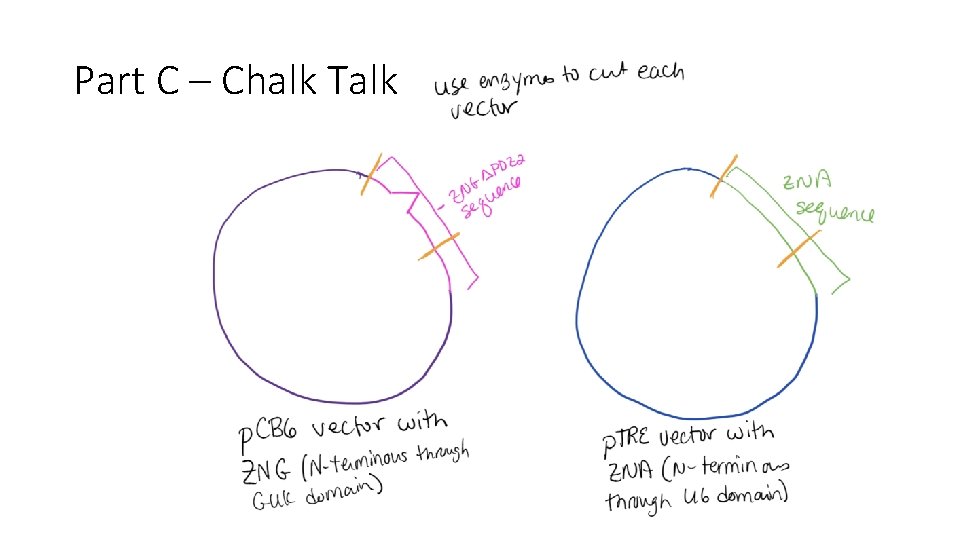
The Study • Express ZO-1 lacking specific binding domains and assess the formation and function of the tight junction. • First we had to create plasmids with ZO-1 altered sequences. • Then we transfected bacteria with the plasmids to confirm plasmid sequence and to generate lots of plasmid • Finally, we transfected MDCK cells lacking ZO-1 and ZO-2 with the plasmids and assessed tight junction structure and function.

This Lab • Walk students through the process of creating plasmids to use for this ZO-1 binding domain function study. • Introduce students to the fact that science doesn’t always work the first time • Introduce students to the fact that science is weird sometimes.

This Workshop – You will be a student • Part A: compare ZO-1 sequence across species and compare ZO sequences • Part B: create a map of domain sequences and restriction enzyme sights within the ZO-1 nucleotide sequence, review the ZO-2 binding domains and binding partners • Part C and D: review process for creating plasmid, determine which plasmids can be used in the experiment • Note: We are working with the N-terminal half of the protein and not the whole protein • If time – look at the published results of our project

Stop when you get to Part C

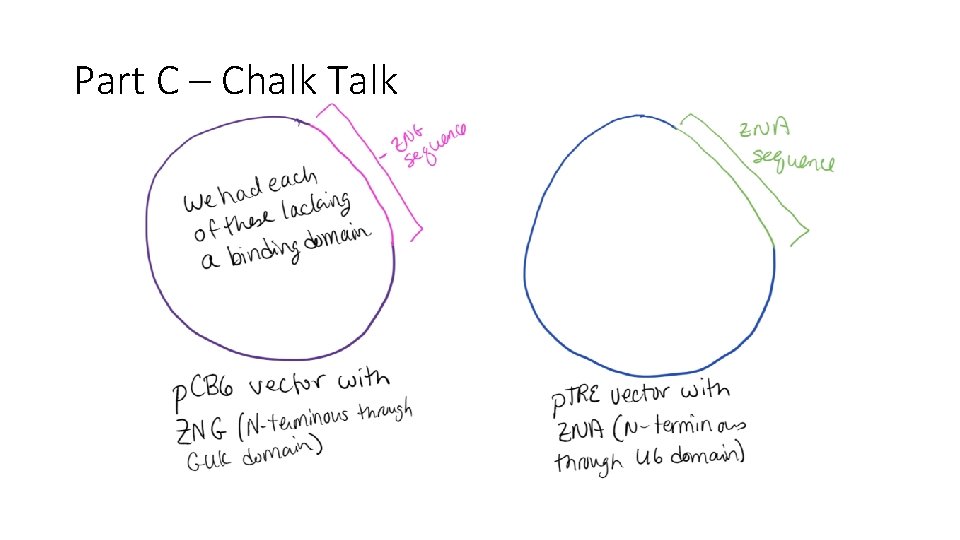
Part C – Chalk Talk

Part C – Chalk Talk

Part C – Chalk Talk
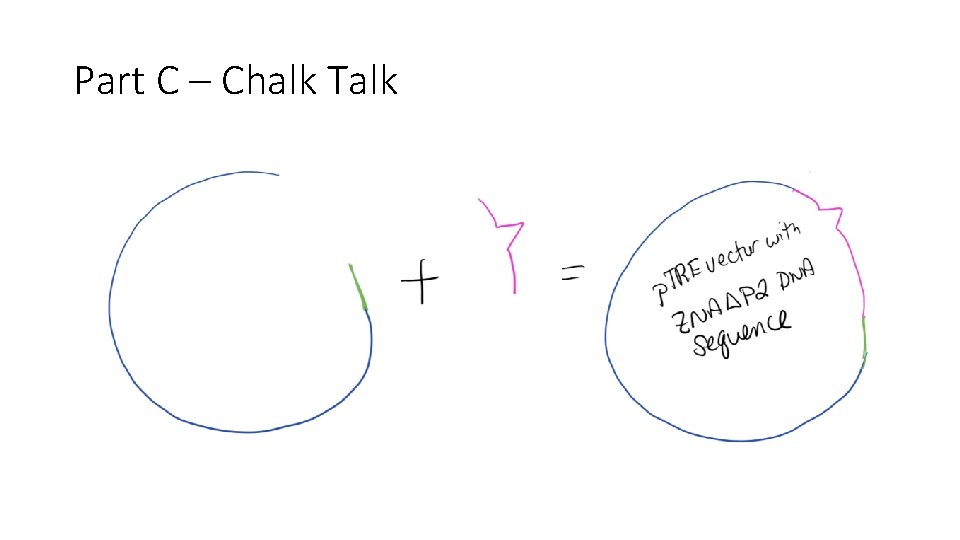
Part C – Chalk Talk