Posttranscriptional events III others 1 Processing of r







- Slides: 27

Post-transcriptional events III: others 1. Processing of r. RNA (eucaryotic and procaryotic) 2. Processing of t. RNA 3. Trans-splicing 4. RNA editing 5. Post-transcriptional control of gene expression

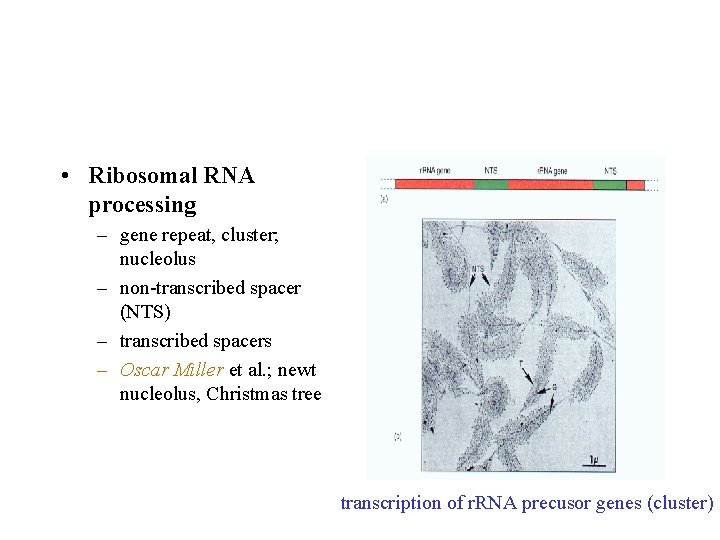
• Ribosomal RNA processing – gene repeat, cluster; nucleolus – non-transcribed spacer (NTS) – transcribed spacers – Oscar Miller et al. ; newt nucleolus, Christmas tree transcription of r. RNA precusor genes (cluster)

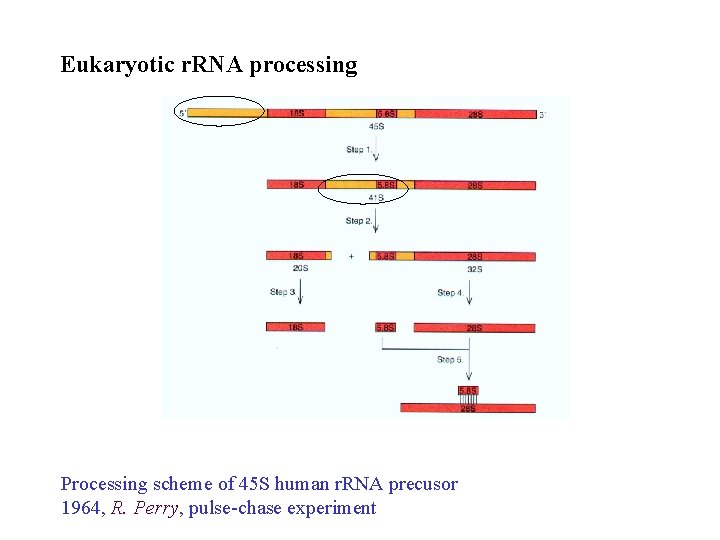
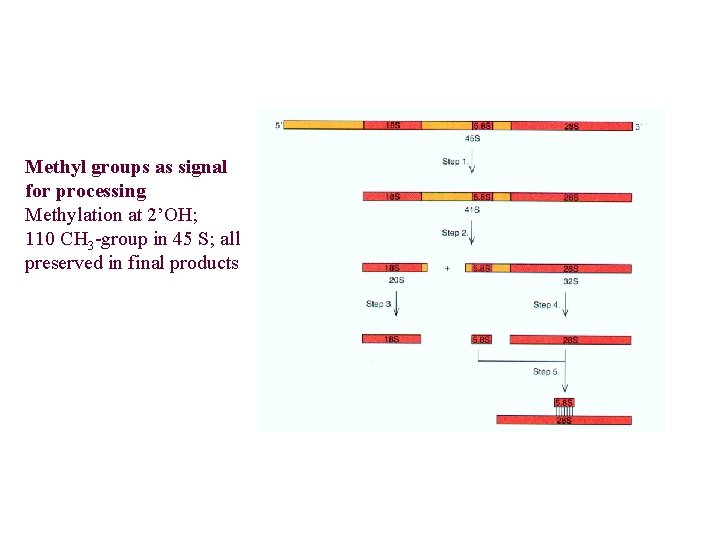
Eukaryotic r. RNA processing Processing scheme of 45 S human r. RNA precusor 1964, R. Perry, pulse-chase experiment

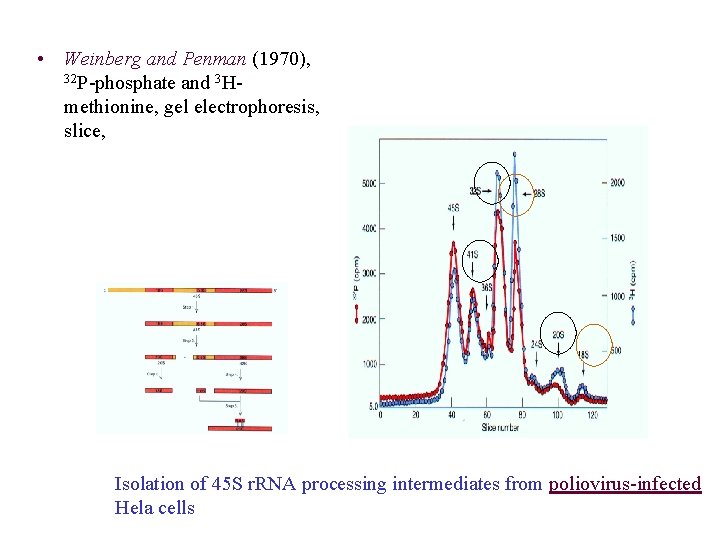
• Weinberg and Penman (1970), 32 P-phosphate and 3 Hmethionine, gel electrophoresis, slice, Isolation of 45 S r. RNA processing intermediates from poliovirus-infected Hela cells

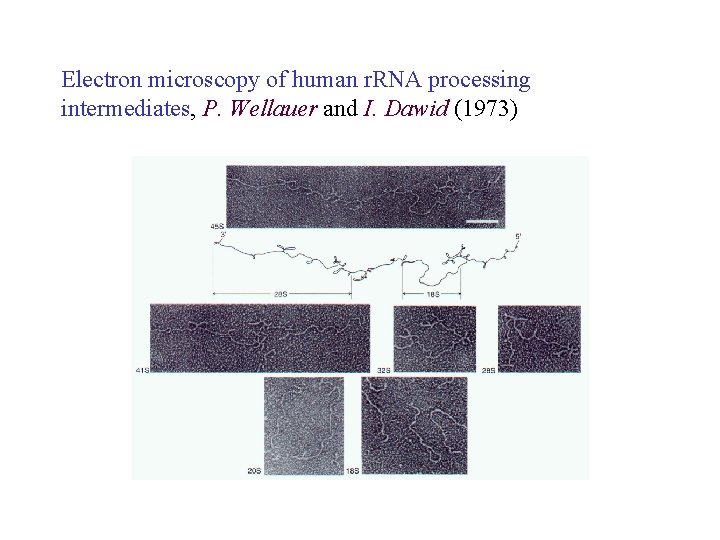
Electron microscopy of human r. RNA processing intermediates, P. Wellauer and I. Dawid (1973)

Methyl groups as signal for processing Methylation at 2’OH; 110 CH 3 -group in 45 S; all preserved in final products

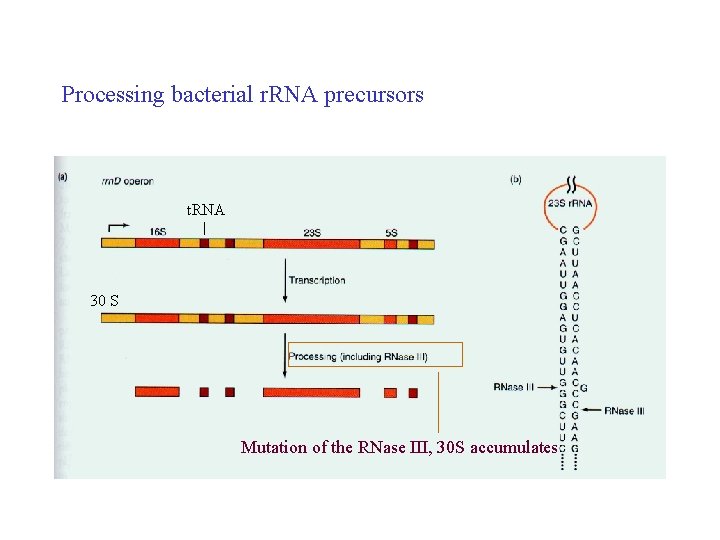
Processing bacterial r. RNA precursors t. RNA 30 S Mutation of the RNase III, 30 S accumulates

• How does the processing apparatus determine what to remove and what to save? – Pattern of methylation, 2’OH – 110 methyl groups in 45 S r. RNA (Hela cells), preserved in mature r. RNA • r. RNAs are made in eukaryotic cells as precursors that must be processed to release the mature r. RNAs. The order of RNAs in precusor is 18 S, 5. 8 S, 28 S in all eukaryotes. • Prokarytoic r. RNA precursors contain t. RNAs as well as all three r. RNAs. The r. RNAs are released from their precuosrs by RNase III and RNase E

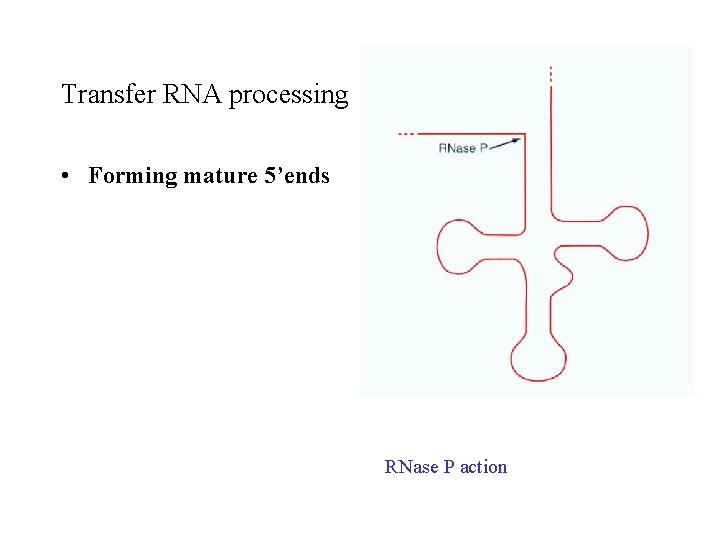
Transfer RNA processing • Forming mature 5’ends RNase P action

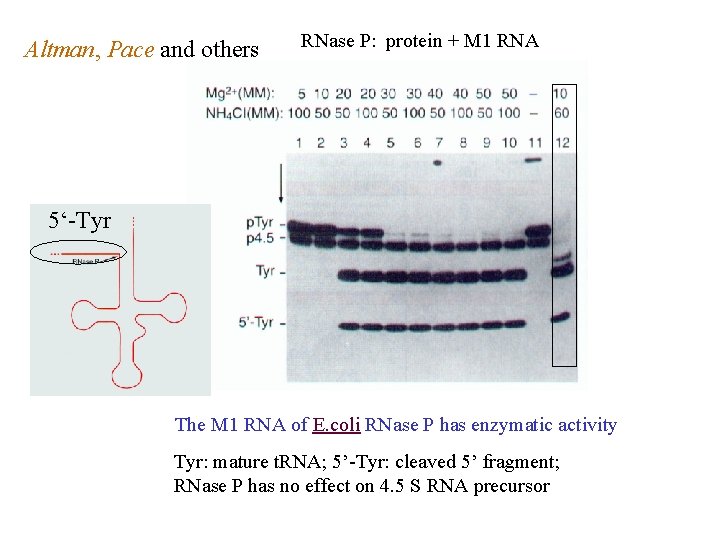
Altman, Pace and others RNase P: protein + M 1 RNA 5‘-Tyr The M 1 RNA of E. coli RNase P has enzymatic activity Tyr: mature t. RNA; 5’-Tyr: cleaved 5’ fragment; RNase P has no effect on 4. 5 S RNA precursor

Eucaryotic RNase P also has an RNA part and it has the enzymatic activity. Spinach chloroplast RNase P appears not to have an RNA part.

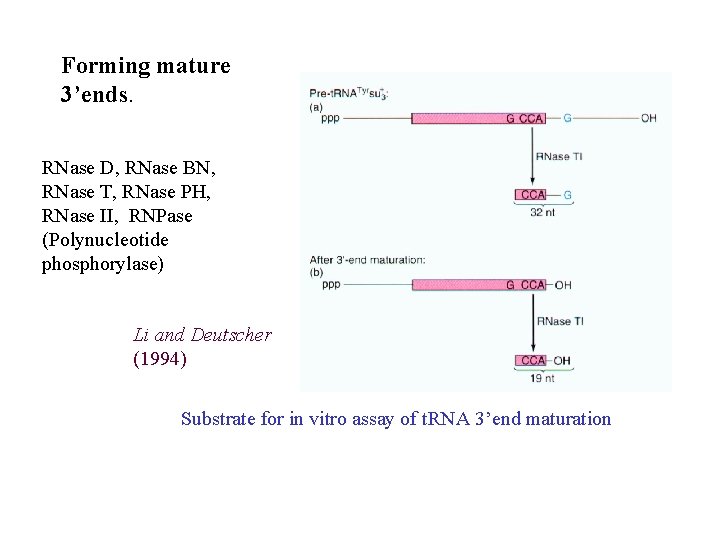
Forming mature 3’ends. RNase D, RNase BN, RNase T, RNase PH, RNase II, RNPase (Polynucleotide phosphorylase) Li and Deutscher (1994) Substrate for in vitro assay of t. RNA 3’end maturation

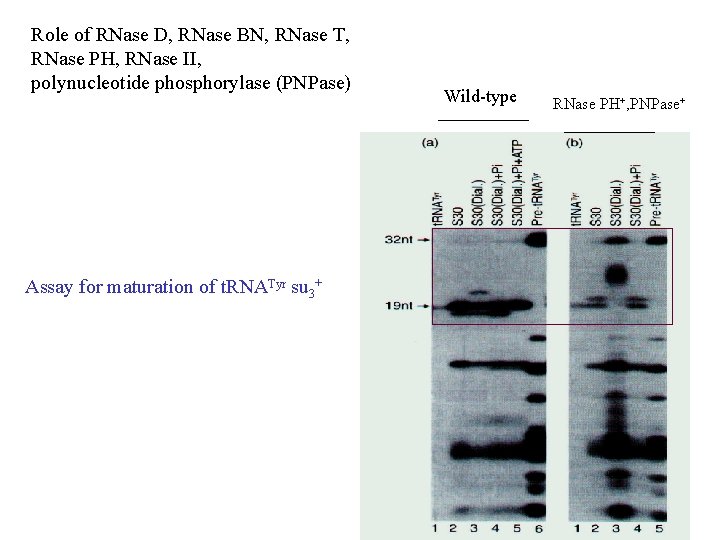
Role of RNase D, RNase BN, RNase T, RNase PH, RNase II, polynucleotide phosphorylase (PNPase) Assay for maturation of t. RNATyr su 3+ Wild-type RNase PH+, PNPase+

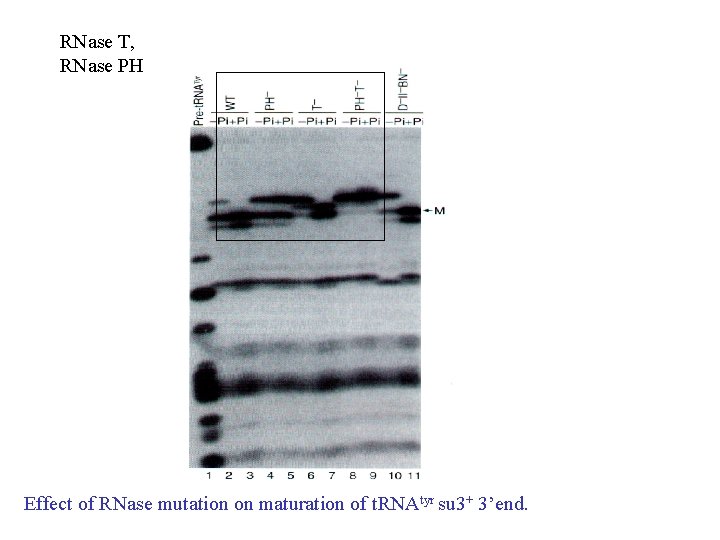
RNase T, RNase PH Effect of RNase mutation on maturation of t. RNAtyr su 3+ 3’end.

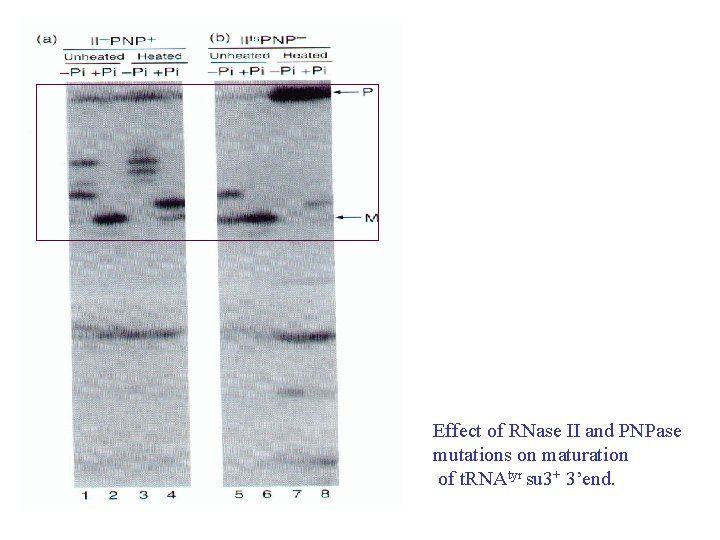
Effect of RNase II and PNPase mutations on maturation of t. RNAtyr su 3+ 3’end.

• RNase II and polynucleotide phosphorylase (PNP) cooperate to remove most of the extra nucleotides at the ends of a t. RNA precursor, but stop at the +2 state with two extra nucleotides remaining; • RNase PH and T are most active in removing the last two nucleotides from the t. RNA with RNase T being the major participants in removing the very last nucleotide.

Trans-splicing vs. Cis-splicing Schistosoma mansoni trypanosome Ascaris lumbricoides Some organisms that trans-splice Euglena

• Piet Borst and coworkers (1982), trypanosome a surface coat protein m. RNA and gene 5’end no match, extra 35 nt in m. RNA. • More m. RNAs discovered to have the extra 35 nt, called the spliced leader (SL) • none of the genes encode the SL • SL is encoded by a gene repeat 200 X, The gene encodes SL plus 100 nt (an intron -like; with 5’ splice sequence)

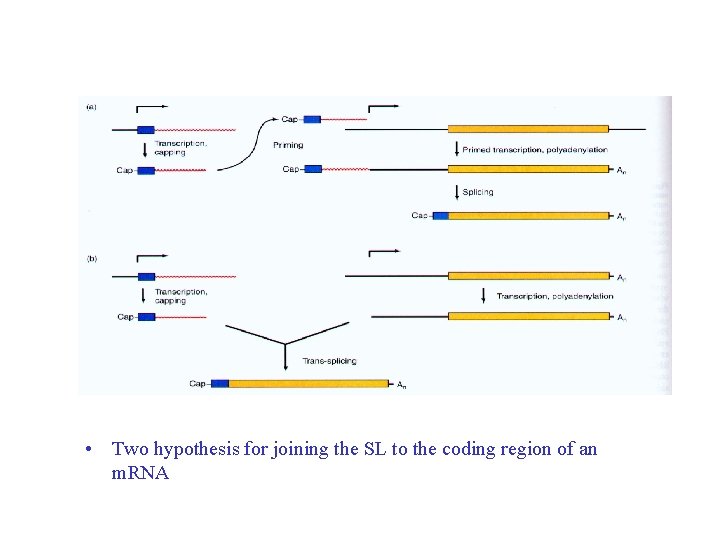
• Two hypothesis for joining the SL to the coding region of an m. RNA

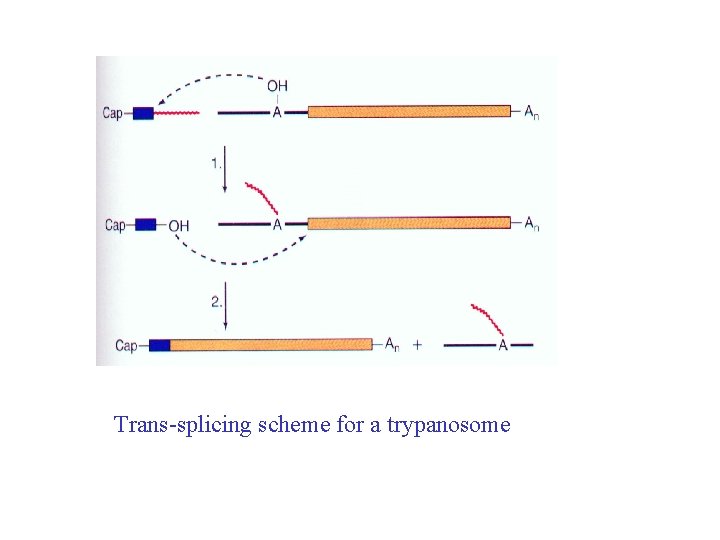
Trans-splicing scheme for a trypanosome

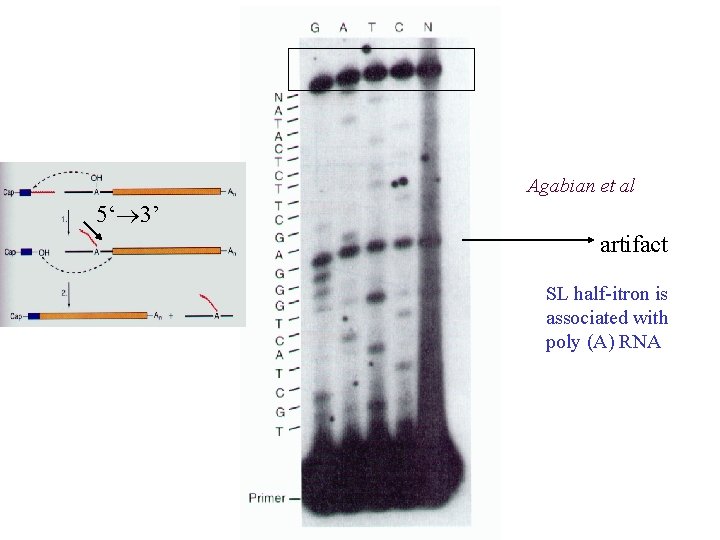
Agabian et al 5‘ 3’ artifact SL half-itron is associated with poly (A) RNA

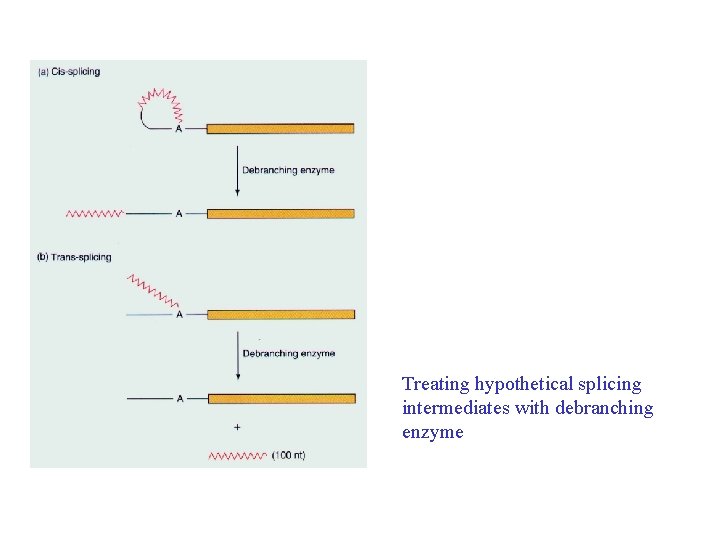
Treating hypothetical splicing intermediates with debranching enzyme

Release of the SL half intron from a larger RNA by debranching enzyme

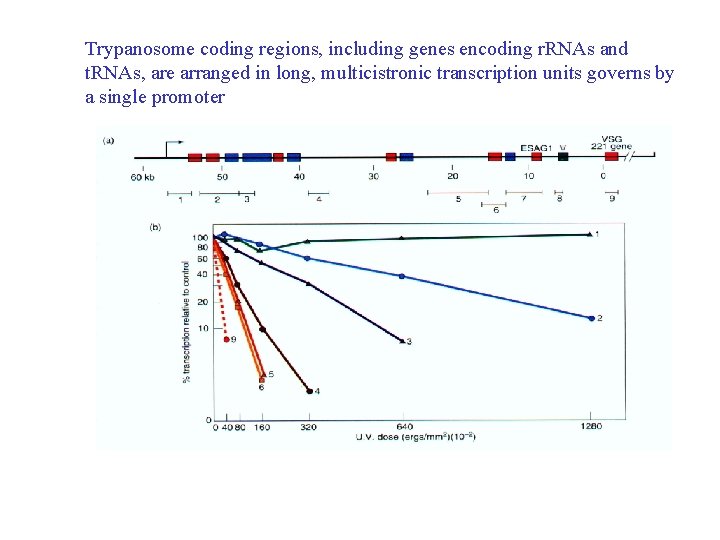
Trypanosome coding regions, including genes encoding r. RNAs and t. RNAs, are arranged in long, multicistronic transcription units governs by a single promoter

Trypanosome m. RNAs are formed by trans-splicing between a short leader exon and any one of many independent coding exon 1. m. RNAs of Trypanosomes have poly(A) tails. 2. However, the genes of the parasites lack of polyadenylation signals.

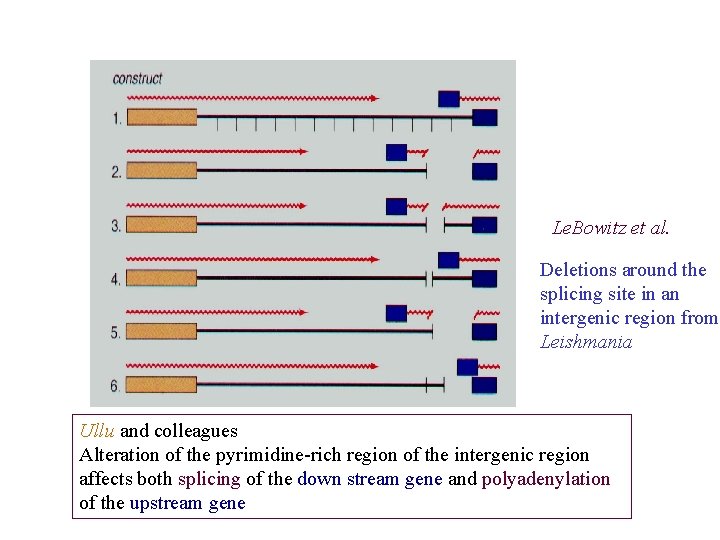
Le. Bowitz et al. Deletions around the splicing site in an intergenic region from Leishmania Ullu and colleagues Alteration of the pyrimidine-rich region of the intergenic region affects both splicing of the down stream gene and polyadenylation of the upstream gene

Summary • Polyadenylation in trypanosomes depends on trans-splicing of the downstream coding region to an SL. • The pyrimidine-rich tract just upstream of the splice site governs both splicing of the downstream gene and polyadenylation of the gene just upstream. • All the genes in a transcription unit are transcribed equally, yet the amounts of the various m. RNAs derived from the transcription unit vary. Control at splicing and polyadenylation level
 Hamlet act iii scene ii
Hamlet act iii scene ii Mutually exclusive vs non mutually exclusive
Mutually exclusive vs non mutually exclusive Top down vs bottom up processing
Top down vs bottom up processing Secondary processed food
Secondary processed food Laplacian filter
Laplacian filter Gloria suarez
Gloria suarez Interactive processing
Interactive processing Point processing in image processing
Point processing in image processing What is point processing in digital image processing
What is point processing in digital image processing Bottom up processing
Bottom up processing Histogram processing in digital image processing
Histogram processing in digital image processing Morphological
Morphological Neighborhood processing
Neighborhood processing Parallel processing vs concurrent processing
Parallel processing vs concurrent processing Bottom up processing example
Bottom up processing example Identifying prepositional phrases
Identifying prepositional phrases Chapter 6 lesson 2 respecting yourself and others
Chapter 6 lesson 2 respecting yourself and others If your actions inspire others
If your actions inspire others Respect each brother
Respect each brother Boyfriend/girlfriend centered paradigm
Boyfriend/girlfriend centered paradigm Motivating yourself and others
Motivating yourself and others Ways to show respect to others
Ways to show respect to others The lives of others music by
The lives of others music by Converse with others
Converse with others Sitxhrm001 coach others in job skills
Sitxhrm001 coach others in job skills Supervising others
Supervising others Overdriving your headlights means
Overdriving your headlights means Perceiving ourselves and others in organizations
Perceiving ourselves and others in organizations