Pediatric Atlases in AFNI Kidding around growing up






- Slides: 21

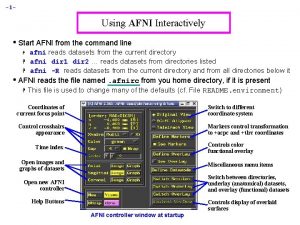
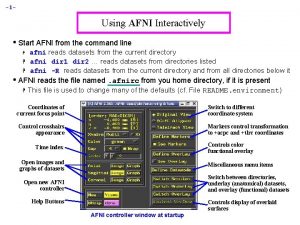
Pediatric Atlases in AFNI "Kidding" around - growing up (and down) with AFNI

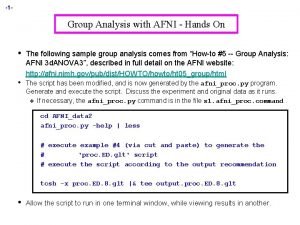
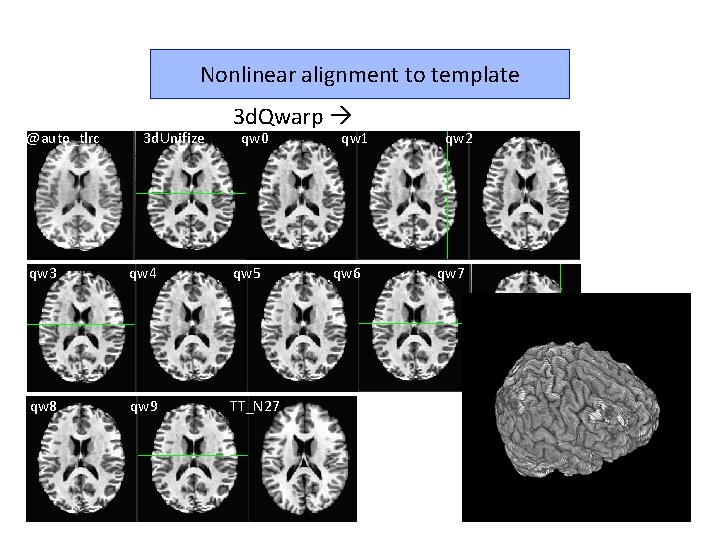
Nonlinear alignment to template @auto_tlrc 3 d. Unifize 3 d. Qwarp qw 0 qw 3 qw 4 qw 5 qw 8 qw 9 TT_N 27 qw 1 qw 6 qw 2 qw 7

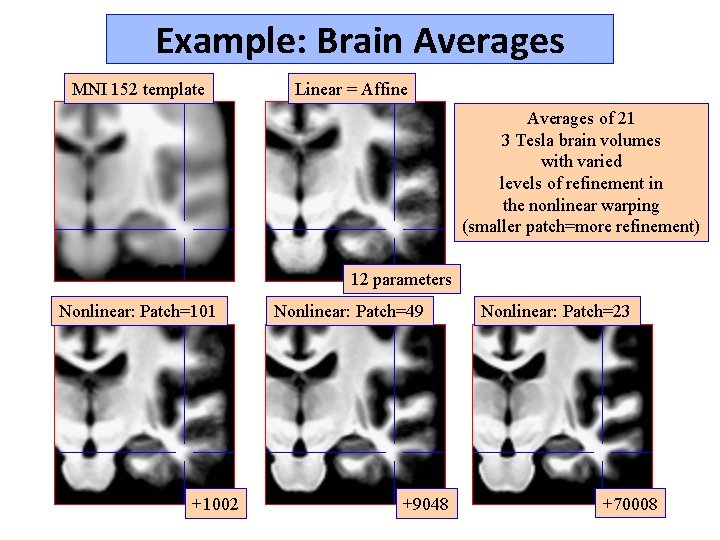
Example: Brain Averages MNI 152 template Linear = Affine Averages of 21 3 Tesla brain volumes with varied levels of refinement in the nonlinear warping (smaller patch=more refinement) 12 parameters Nonlinear: Patch=101 +1002 Nonlinear: Patch=49 +9048 Nonlinear: Patch=23 +70008

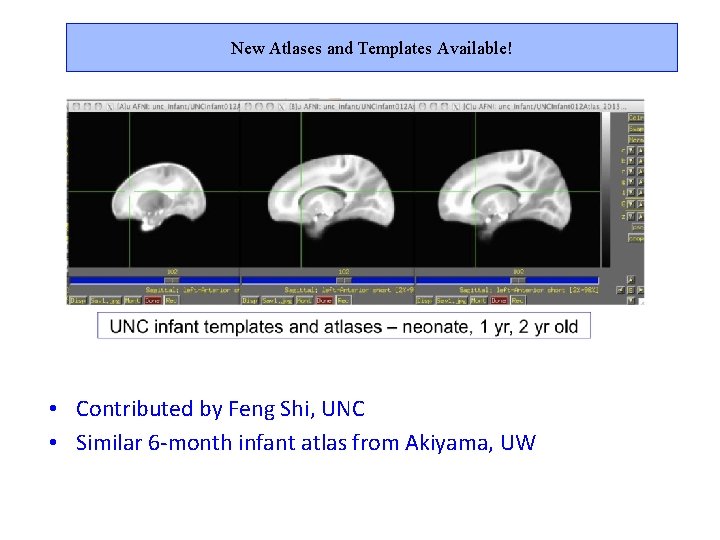
New Atlases and Templates Available! • Contributed by Feng Shi, UNC • Similar 6 -month infant atlas from Akiyama, UW

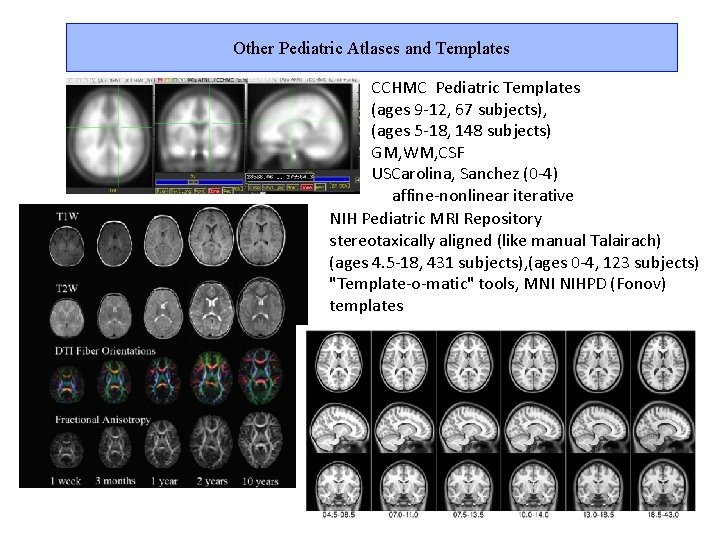
Other Pediatric Atlases and Templates CCHMC Pediatric Templates (ages 9 -12, 67 subjects), (ages 5 -18, 148 subjects) GM, WM, CSF USCarolina, Sanchez (0 -4) affine-nonlinear iterative NIH Pediatric MRI Repository stereotaxically aligned (like manual Talairach) (ages 4. 5 -18, 431 subjects), (ages 0 -4, 123 subjects) "Template-o-matic" tools, MNI NIHPD (Fonov) templates

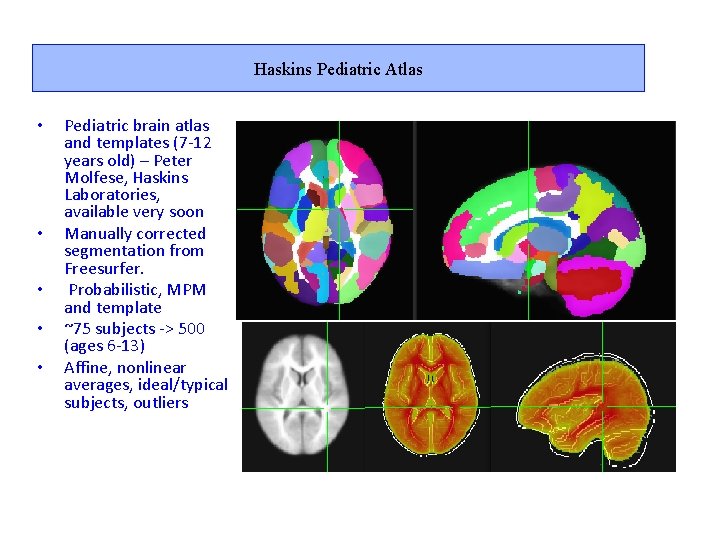
Haskins Pediatric Atlas • • • Pediatric brain atlas and templates (7 -12 years old) – Peter Molfese, Haskins Laboratories, available very soon Manually corrected segmentation from Freesurfer. Probabilistic, MPM and template ~75 subjects -> 500 (ages 6 -13) Affine, nonlinear averages, ideal/typical subjects, outliers

Haskins Pediatric Atlas – "Atlasing" options 1. Affine group – unifize, iterative affine alignment and averaging 2. Nonlinear group – unifize, affine align to 1, iterative nonlinear in varying neighborhood sizes and averaging 3. Nonlinear individual – "typical" individual 4. Nonlinear group II – nonlinear align all to 3 and average 5. Nonlinear group III –iterative. Use 3 as starting template and proceed as in 2. 6. Nonlinear individual II. "Ideal" individual subject. Align typical from 3 to the group template from 5.

Haskins Pediatric Atlas – Affine template building 1. Affine to MNI 152 (never actually use this result) 2. Rigid Equivalent to MNI 152 (useful byproduct of step 1) 3. Mean (2) 4. @auto_tlrc -base (3) -input originals 5. Mean (4) 6. @auto_tlrc -base (5) -input originals 7. Mean (6) 8. etc. Making the atlas (segmentations) 1. Transform the segmentations similarly as subject to template 2. Average each transformed region separately to find probability of that voxel containing that region (>0. 05) 3. Make maximum probability maps by finding region with maximum probability (smooth with 3 d. Local. Histog)

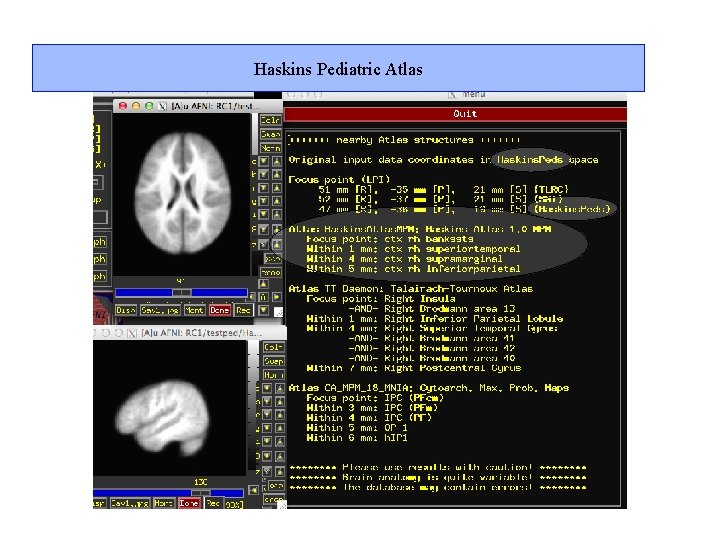
Haskins Pediatric Atlas

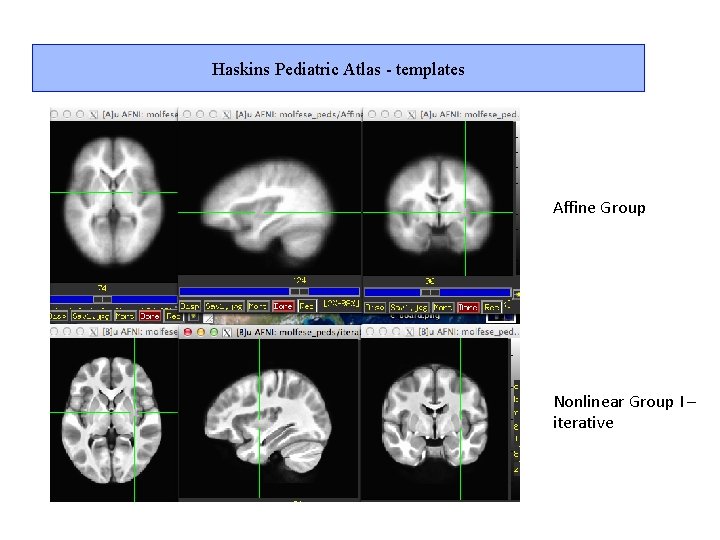
Haskins Pediatric Atlas - templates Affine Group Nonlinear Group I – iterative

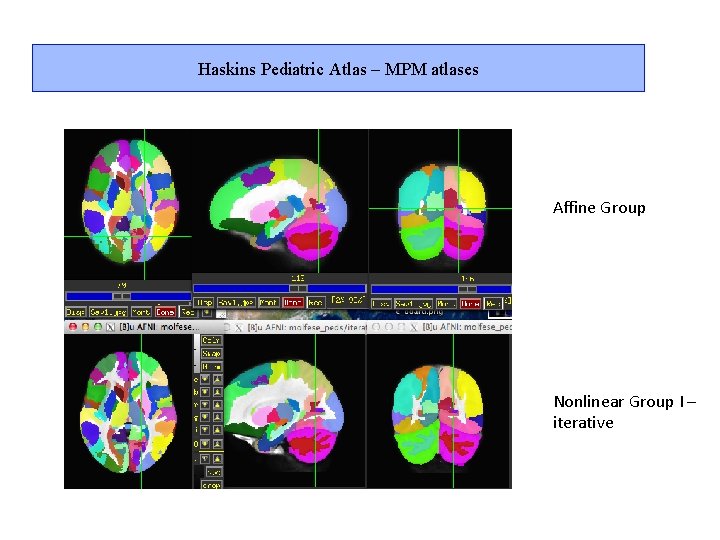
Haskins Pediatric Atlas – MPM atlases Affine Group Nonlinear Group I – iterative

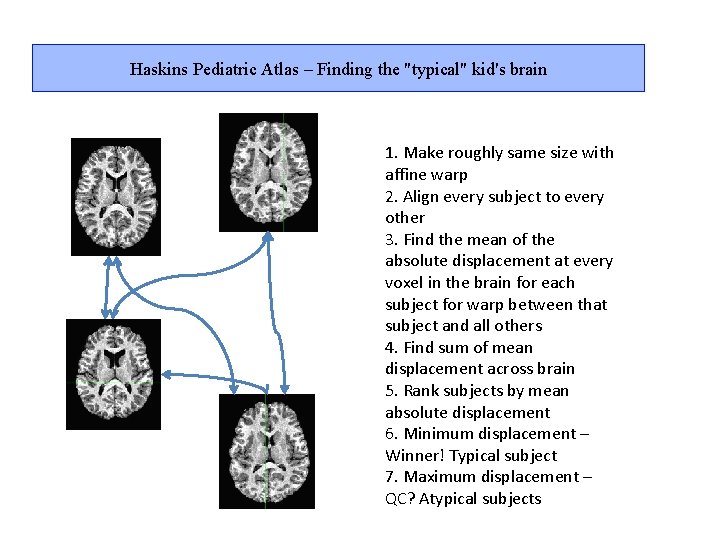
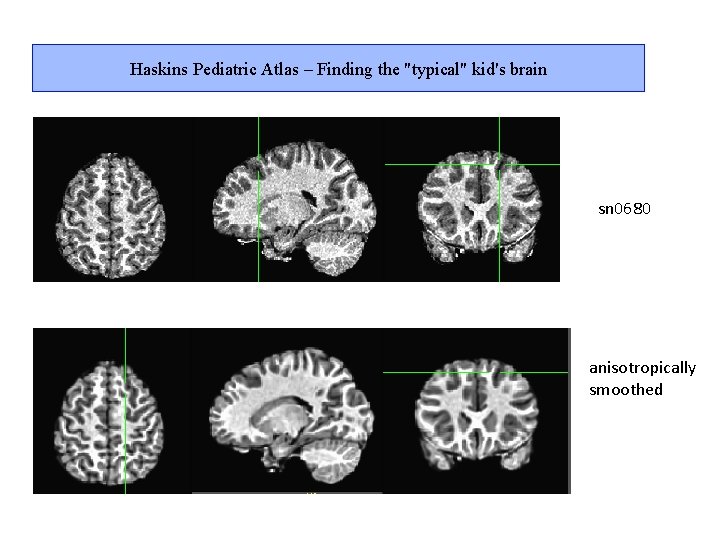
Haskins Pediatric Atlas – Finding the "typical" kid's brain 1. Make roughly same size with affine warp 2. Align every subject to every other 3. Find the mean of the absolute displacement at every voxel in the brain for each subject for warp between that subject and all others 4. Find sum of mean displacement across brain 5. Rank subjects by mean absolute displacement 6. Minimum displacement – Winner! Typical subject 7. Maximum displacement – QC? Atypical subjects

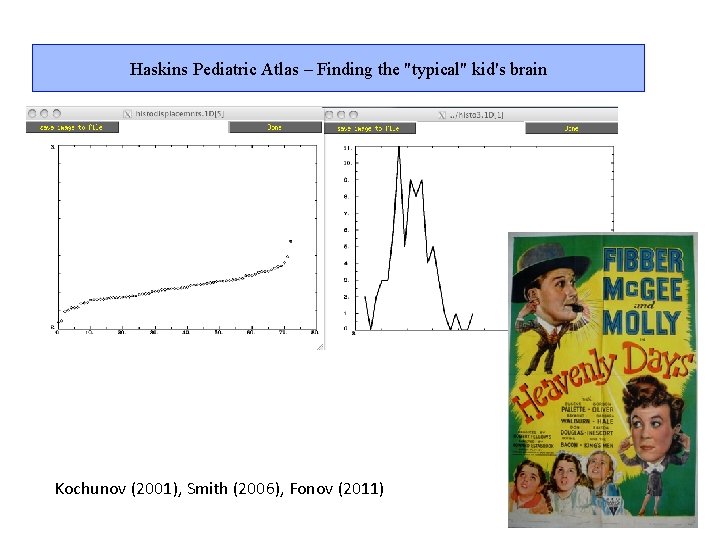
Haskins Pediatric Atlas – Finding the "typical" kid's brain Kochunov (2001), Smith (2006), Fonov (2011)

Haskins Pediatric Atlas – Finding the "typical" kid's brain sn 0680 anisotropically smoothed

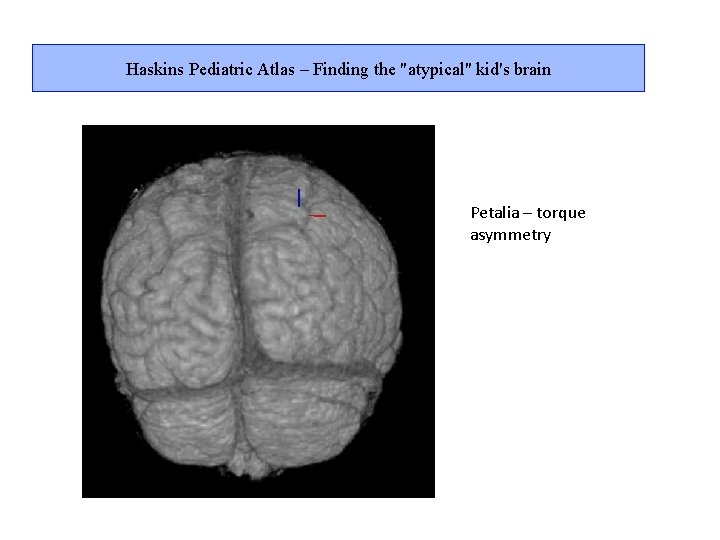
Haskins Pediatric Atlas – Finding the "atypical" kid's brain Petalia – torque asymmetry

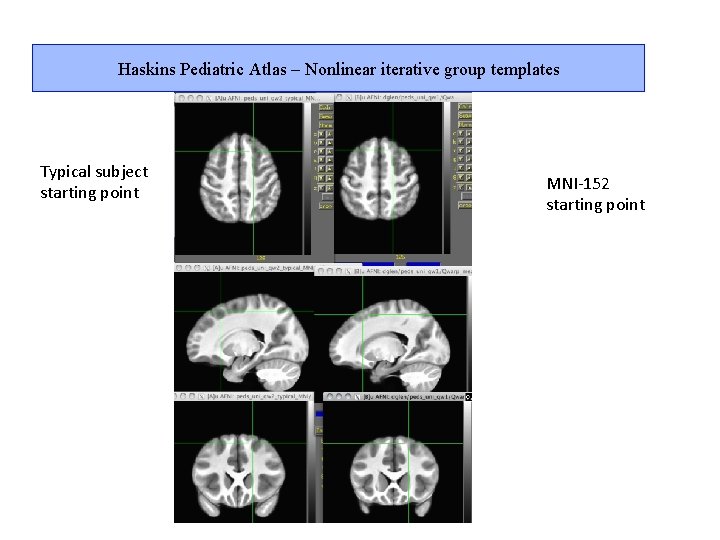
Haskins Pediatric Atlas – Nonlinear iterative group templates Typical subject starting point MNI-152 starting point

Haskins Pediatric Atlas – Decisions, Decisions Regions – some to leave in, some to take out. Small regions can get wiped out. Large white matter regions from Freesurfer removed Small variable regions (about 5) disappear Consider probability map atlases and neighboring ROIs Histogram smoothing Individual versus Group template – bias / detail Age range – split into smaller ranges as group gets larger. How many required for stable template? Too few, too many?

Haskins Pediatric Atlas – Thanks Peter Molfese, Haskins -> U. Conn. Laura Mesite, Haskins -> Harvard Robert Cox - NIH

Bonus Extra Features

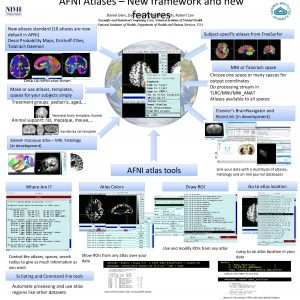
Web-based atlases Neuro. Synth, Linkr. Brain, Brain. Map. org, Allen Brain, Elsevier Brain. Navigator, . . . set AFNI_WEBBY_WAMI to YES and AFNI_NEUROSYNTH to YES

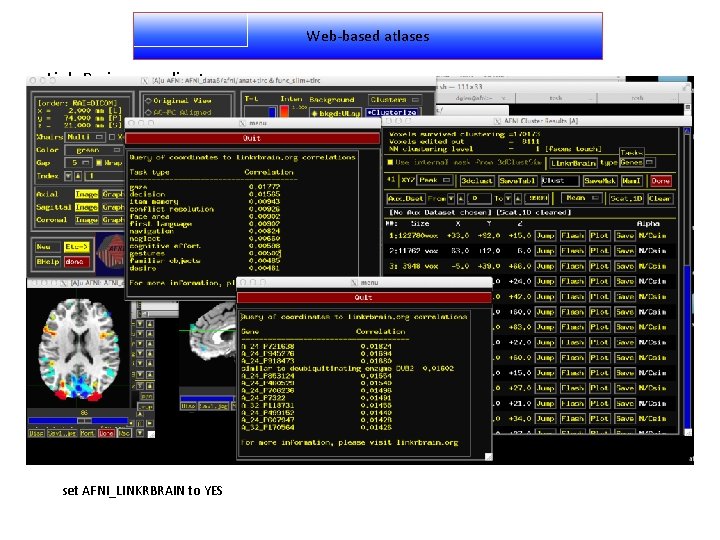
Web-based atlases Linkr. Brain - coordinates set AFNI_LINKRBRAIN to YES
 Packing sprite atlases
Packing sprite atlases Just kidding disney
Just kidding disney Just kidding star
Just kidding star Afni fmri
Afni fmri Afni fmri
Afni fmri Fft
Fft Afni group analysis
Afni group analysis Erp afni
Erp afni Does afni drug test
Does afni drug test Niml
Niml Afni group analysis
Afni group analysis Afni group analysis
Afni group analysis Afni group analysis
Afni group analysis Afni dimon
Afni dimon Martin luther king of hinduism
Martin luther king of hinduism What goes around comes around examples
What goes around comes around examples Facts about manifest destiny
Facts about manifest destiny Hawaii hardiness zone
Hawaii hardiness zone Prayer for grade 3
Prayer for grade 3 Why do cells divide instead of growing larger
Why do cells divide instead of growing larger Growing and changing
Growing and changing Growing knowing
Growing knowing