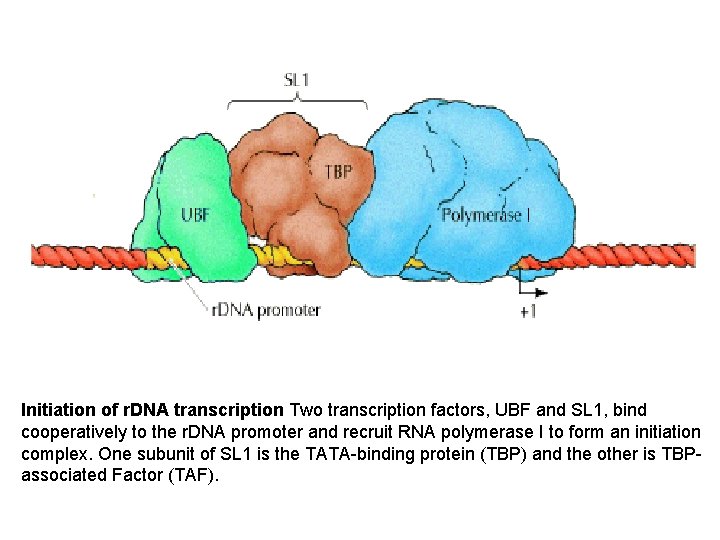
Initiation of r DNA transcription Two transcription factors


- Slides: 12

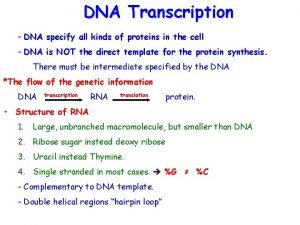
Initiation of r. DNA transcription Two transcription factors, UBF and SL 1, bind cooperatively to the r. DNA promoter and recruit RNA polymerase I to form an initiation complex. One subunit of SL 1 is the TATA-binding protein (TBP) and the other is TBP- associated Factor (TAF).

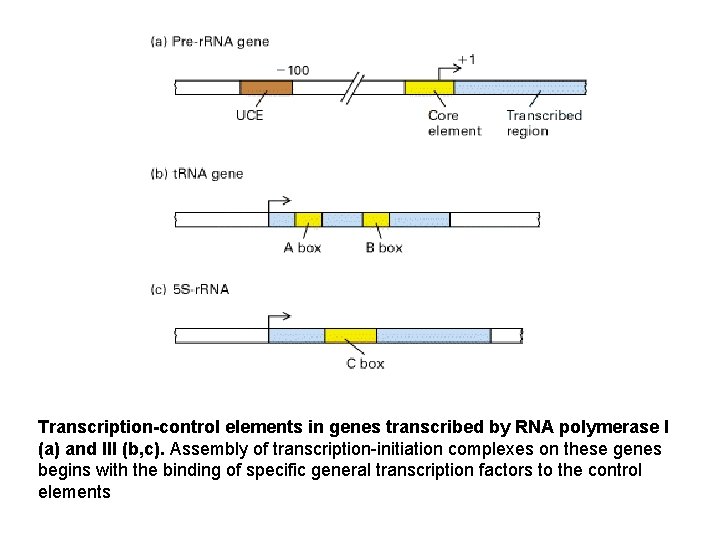
Transcription-control elements in genes transcribed by RNA polymerase I (a) and III (b, c). Assembly of transcription-initiation complexes on these genes begins with the binding of specific general transcription factors to the control elements

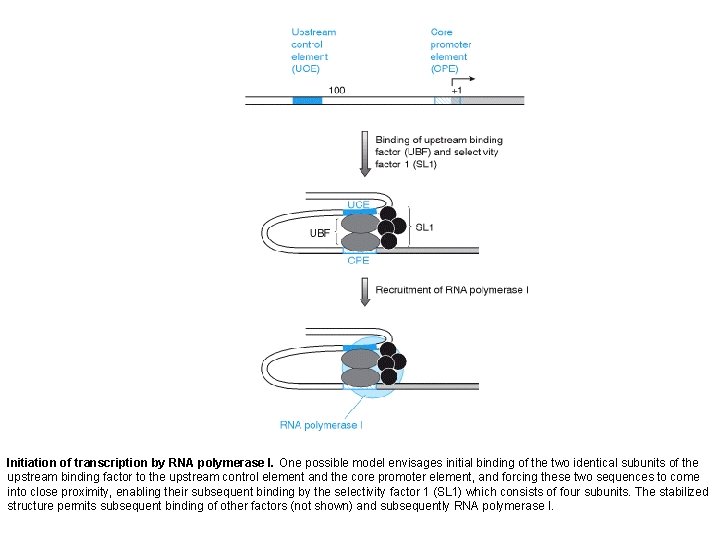
Initiation of transcription by RNA polymerase I. One possible model envisages initial binding of the two identical subunits of the upstream binding factor to the upstream control element and the core promoter element, and forcing these two sequences to come into close proximity, enabling their subsequent binding by the selectivity factor 1 (SL 1) which consists of four subunits. The stabilized structure permits subsequent binding of other factors (not shown) and subsequently RNA polymerase I.

Examples of chemical modifications occurring with nucleotides in r. RNA and t. RNA Modification Details Example Methylation Addition of one or more -CH 3 groups to the base or sugar Methylation of guanosine gives 7 -methylguanosine Deamination Removal of an amino (-NH 2) group from the base Deamination of adenosine gives inosine Sulfur substitution Replacement of oxygen with sulfur 4 -Thiouridine Base isomerization Changing the positions of atoms in the ring component of the base Isomerization of uridine gives pseudouridine Double-bond saturation Converting a double bond to a single bond Double bond saturation converts uridine to dihydrouridine

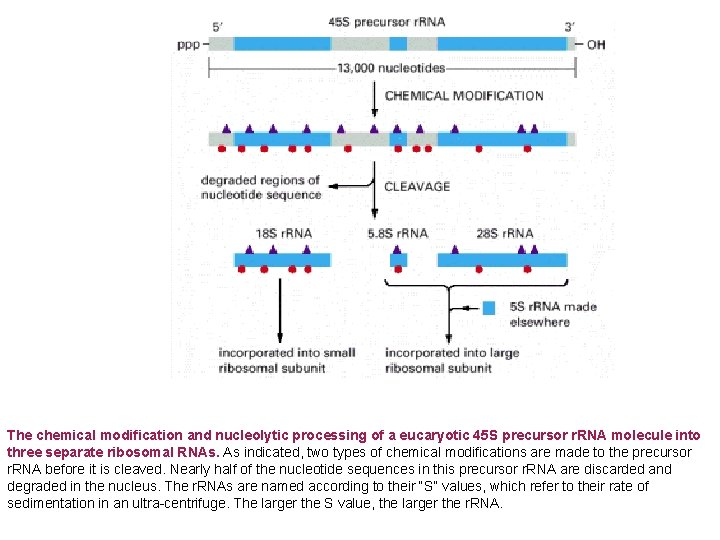
The chemical modification and nucleolytic processing of a eucaryotic 45 S precursor r. RNA molecule into three separate ribosomal RNAs. As indicated, two types of chemical modifications are made to the precursor r. RNA before it is cleaved. Nearly half of the nucleotide sequences in this precursor r. RNA are discarded and degraded in the nucleus. The r. RNAs are named according to their “S” values, which refer to their rate of sedimentation in an ultra-centrifuge. The larger the S value, the larger the r. RNA.

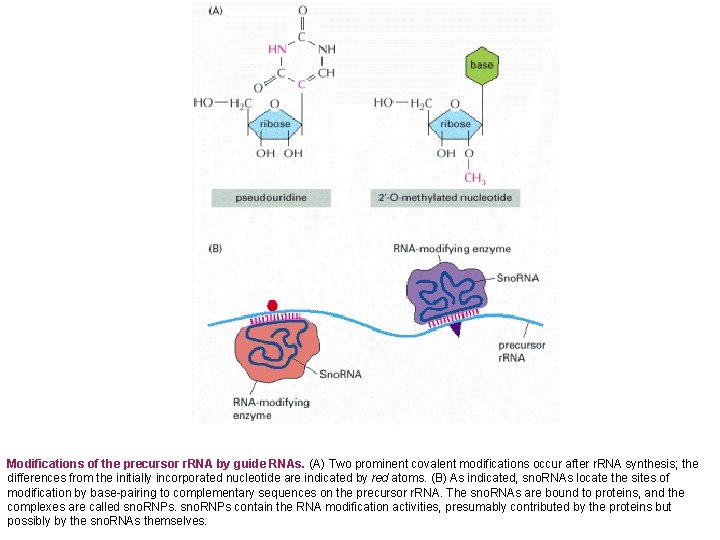
Modifications of the precursor r. RNA by guide RNAs. (A) Two prominent covalent modifications occur after r. RNA synthesis; the differences from the initially incorporated nucleotide are indicated by red atoms. (B) As indicated, sno. RNAs locate the sites of modification by base-pairing to complementary sequences on the precursor r. RNA. The sno. RNAs are bound to proteins, and the complexes are called sno. RNPs contain the RNA modification activities, presumably contributed by the proteins but possibly by the sno. RNAs themselves.

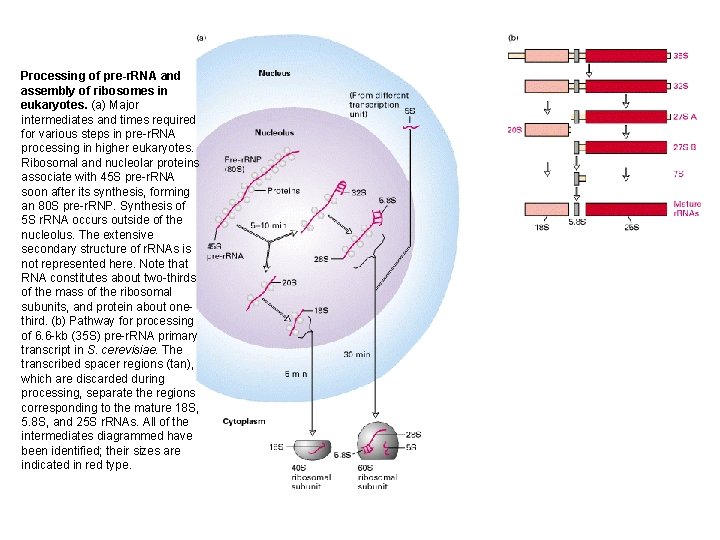
Processing of pre-r. RNA and assembly of ribosomes in eukaryotes. (a) Major intermediates and times required for various steps in pre-r. RNA processing in higher eukaryotes. Ribosomal and nucleolar proteins associate with 45 S pre-r. RNA soon after its synthesis, forming an 80 S pre-r. RNP. Synthesis of 5 S r. RNA occurs outside of the nucleolus. The extensive secondary structure of r. RNAs is not represented here. Note that RNA constitutes about two-thirds of the mass of the ribosomal subunits, and protein about onethird. (b) Pathway for processing of 6. 6 -kb (35 S) pre-r. RNA primary transcript in S. cerevisiae. The transcribed spacer regions (tan), which are discarded during processing, separate the regions corresponding to the mature 18 S, 5. 8 S, and 25 S r. RNAs. All of the intermediates diagrammed have been identified; their sizes are indicated in red type.

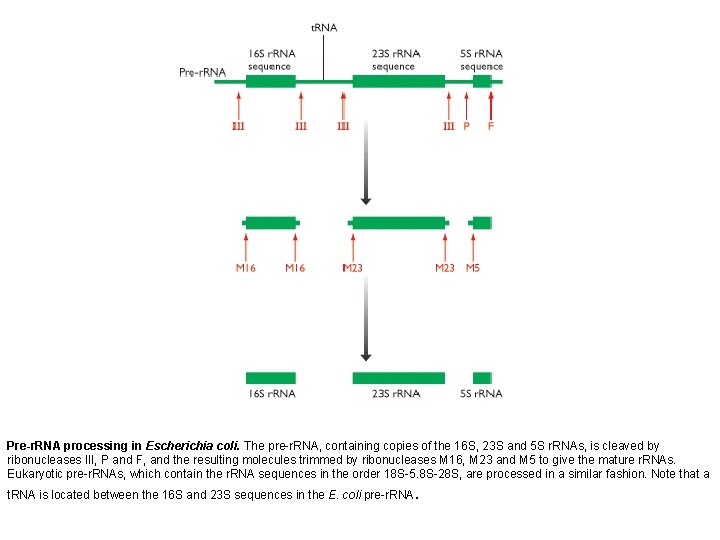
Pre-r. RNA processing in Escherichia coli. The pre-r. RNA, containing copies of the 16 S, 23 S and 5 S r. RNAs, is cleaved by ribonucleases III, P and F, and the resulting molecules trimmed by ribonucleases M 16, M 23 and M 5 to give the mature r. RNAs. Eukaryotic pre-r. RNAs, which contain the r. RNA sequences in the order 18 S-5. 8 S-28 S, are processed in a similar fashion. Note that a t. RNA is located between the 16 S and 23 S sequences in the E. coli pre-r. RNA.

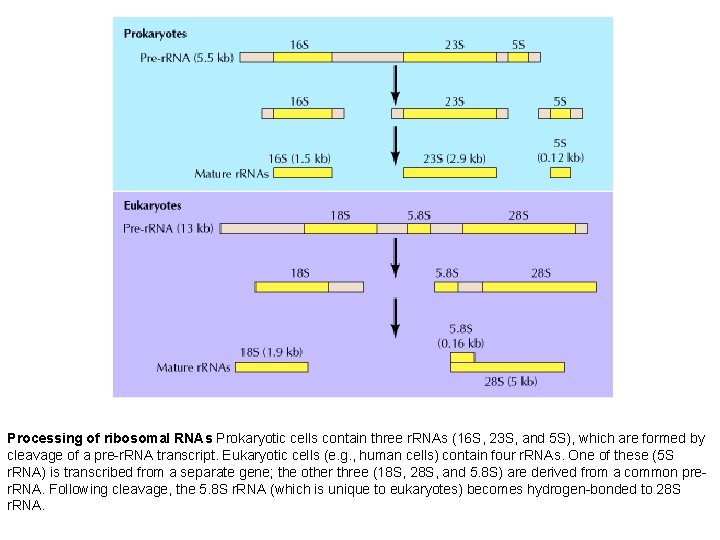
Processing of ribosomal RNAs Prokaryotic cells contain three r. RNAs (16 S, 23 S, and 5 S), which are formed by cleavage of a pre-r. RNA transcript. Eukaryotic cells (e. g. , human cells) contain four r. RNAs. One of these (5 S r. RNA) is transcribed from a separate gene; the other three (18 S, 28 S, and 5. 8 S) are derived from a common prer. RNA. Following cleavage, the 5. 8 S r. RNA (which is unique to eukaryotes) becomes hydrogen-bonded to 28 S r. RNA.

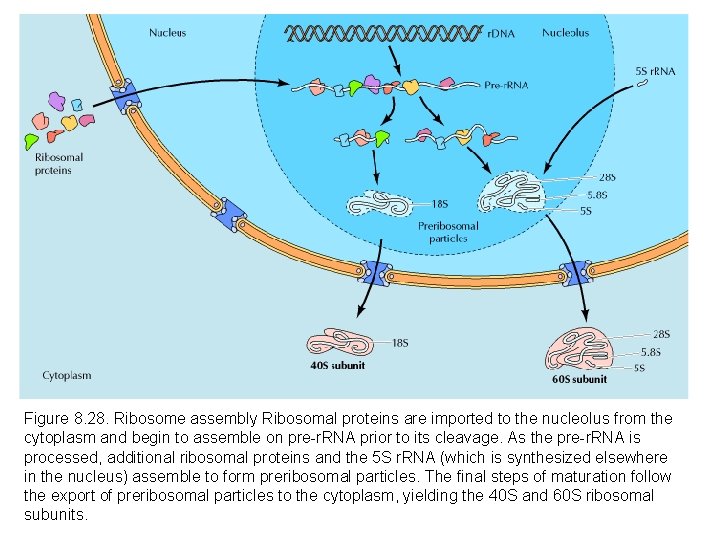
Figure 8. 28. Ribosome assembly Ribosomal proteins are imported to the nucleolus from the cytoplasm and begin to assemble on pre-r. RNA prior to its cleavage. As the pre-r. RNA is processed, additional ribosomal proteins and the 5 S r. RNA (which is synthesized elsewhere in the nucleus) assemble to form preribosomal particles. The final steps of maturation follow the export of preribosomal particles to the cytoplasm, yielding the 40 S and 60 S ribosomal subunits.


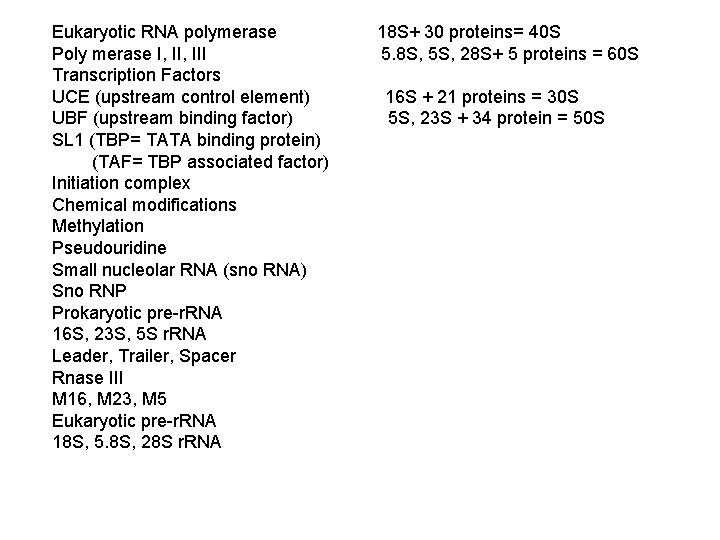
Eukaryotic RNA polymerase 18 S+ 30 proteins= 40 S Poly merase I, III 5. 8 S, 5 S, 28 S+ 5 proteins = 60 S Transcription Factors UCE (upstream control element) 16 S + 21 proteins = 30 S UBF (upstream binding factor) 5 S, 23 S + 34 protein = 50 S SL 1 (TBP= TATA binding protein) (TAF= TBP associated factor) Initiation complex Chemical modifications Methylation Pseudouridine Small nucleolar RNA (sno RNA) Sno RNP Prokaryotic pre-r. RNA 16 S, 23 S, 5 S r. RNA Leader, Trailer, Spacer Rnase III M 16, M 23, M 5 Eukaryotic pre-r. RNA 18 S, 5. 8 S, 28 S r. RNA
 Rho independent termination
Rho independent termination Transcription and translation coloring
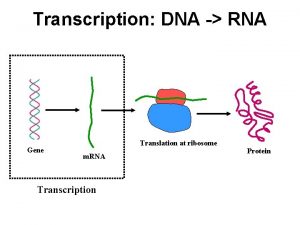
Transcription and translation coloring Dna to rna transcription
Dna to rna transcription Dna transcription and translation
Dna transcription and translation Acids and bases venn diagram
Acids and bases venn diagram Dna replication transcription and translation
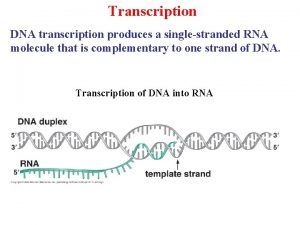
Dna replication transcription and translation Dna transcription
Dna transcription Dna and transcription tutorial
Dna and transcription tutorial Dna to rna transcription
Dna to rna transcription Dna transcription
Dna transcription Dna transcription
Dna transcription Dna transcription
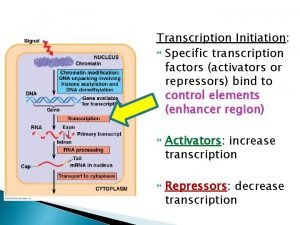
Dna transcription Activation domain of transcription factors
Activation domain of transcription factors