ILSI HESI Genotoxic Mechanisms Cytotoxicity Profiles Realtime PCR




- Slides: 13

ILSI / HESI - Genotoxic Mechanisms Cytotoxicity Profiles & Real-time PCR data from Tanabe (Taxol, Cisplatin in TK 6) Chinami ARUGA Exploratory Toxicology and DMPK Research Laboratories, Tanabe Seiyaku Co. , Ltd. 22 -Jan-2007

Study design Human TK 6 cells Chemicals: low, mid and high doses + time-matched controls Cisplatin Taxol (0. 1, 1 and 10 ug/m. L) (0. 01, 0. 03 and 0. 1 ug/m. L) Treatment 4 hr-treatment (4 hr) 4 hr-treatment + 3 hr-recovery (7 hr) 4 hr-treatment + 20 hr-recovery (24 hr) 2 independent biological replicates x 2 technical replicates Micronucleus observation in 24 hr RT-PCR (47+1 genes) ABI 7900 Low density arrays (cards) Housekeeping gene: 18 S Automatic threshold used on SDS v 2. 2 software partly reanalyzed using a manually fixed threshold DDCt method using the respective control at each time point

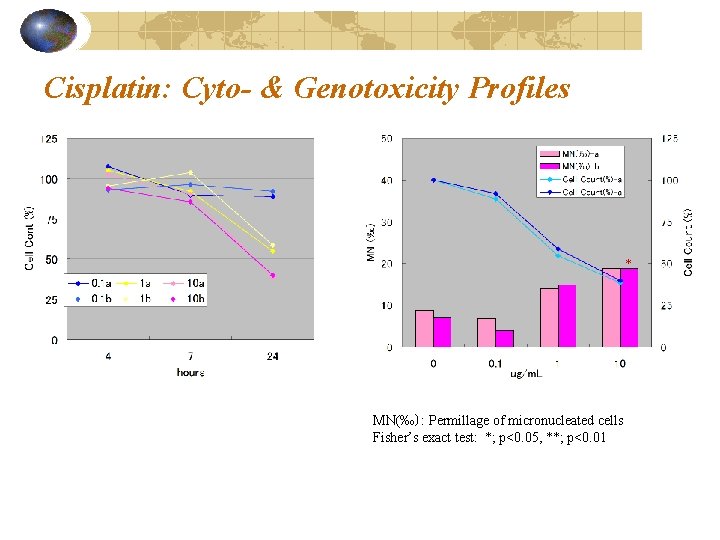
Cisplatin: Cyto- & Genotoxicity Profiles * MN(‰): Permillage of micronucleated cells Fisher’s exact test: *; p<0. 05, **; p<0. 01

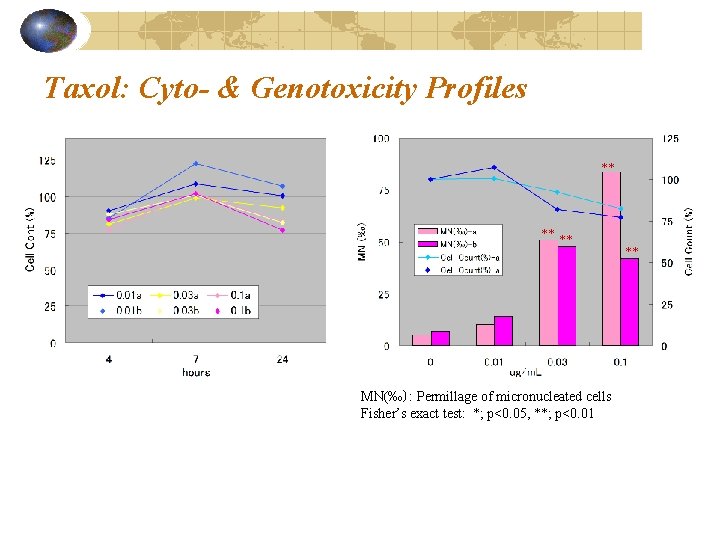
Taxol: Cyto- & Genotoxicity Profiles ** ** ** MN(‰): Permillage of micronucleated cells Fisher’s exact test: *; p<0. 05, **; p<0. 01 **

Cisplatin: PCR data x -8 x -4 x – 1. 5 x 4 x 8

Taxol: PCR data x -8 x -4 x – 1. 5 x 4 x 8

Up-regulation Ten genes were up-regulated by both direct and indirect genotoxicant. Four genes were up-regulated only by direct genotoxicant. Two genes were up-regulated only by indirect genotoxicant. x -8 x -4 x – 1. 5 x 4 x 8

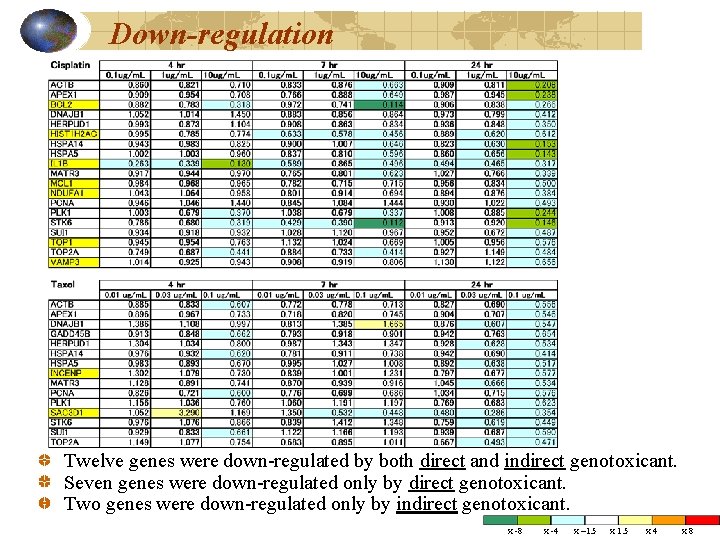
Down-regulation Twelve genes were down-regulated by both direct and indirect genotoxicant. Seven genes were down-regulated only by direct genotoxicant. Two genes were down-regulated only by indirect genotoxicant. x -8 x -4 x – 1. 5 x 4 x 8

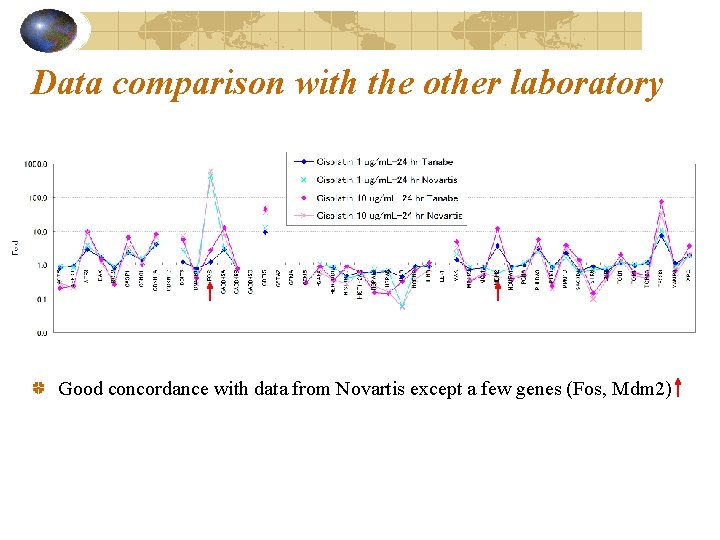
Data comparison with the other laboratory Good concordance with data from Novartis except a few genes (Fos, Mdm 2)

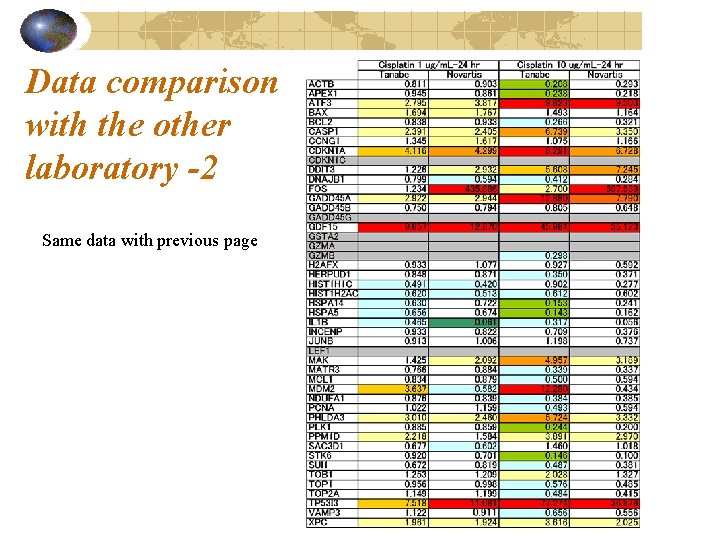
Data comparison with the other laboratory -2 Same data with previous page

Outlines Genes not detected GSTA 2, GZMA, GZMB, LEF 1, CDKN 1 C, GADD 45 C. Dose & Time dependency Taxol: Change of gene expression was moderate even in severe genotoxicity concentration. 22 genes were up- or down-regulated by both chemicals. GDF 15, TP 53 I 3 were prominently up-regulated by both chemicals. 15 genes were specifically up- or down-regulated by each chemical. Good concordance with data from the other laboratory.

Next Step Etoposide, Na. Cl study in TK 6 Search for the discriminating genes Comparison with data from the other companies

Acknowledgment Tanabe, Exploratory Toxicology Group Hideko Yamazaki: Cell Culture and Cyto-&Genotox , RT-PCR and Analysis Naoko Murakami: RT-PCR Hisako Fujimura Wataru Toriumi
 Hesi lonestar
Hesi lonestar Cscc hesi
Cscc hesi Lonestar lvn to rn transition
Lonestar lvn to rn transition Realtime aps software
Realtime aps software Frankfurt realtime
Frankfurt realtime Ams realtime weather maps central
Ams realtime weather maps central Cac realtime
Cac realtime Forrester wave real time interaction management
Forrester wave real time interaction management Realtime iep
Realtime iep Realtime optimization
Realtime optimization Realtime big data
Realtime big data Real time characteristics of embedded operating systems
Real time characteristics of embedded operating systems Realtime it
Realtime it Realtime database push
Realtime database push