GRCh 37 GM 12878 Segway segmentation comparison with





- Slides: 12

GRCh 37 GM 12878 Segway segmentation & comparison with K 562 Steven Wilder EBI http: //encodewiki. ucsc. edu/Encode. DCC /index. php/Integration_Vignette_S 04 1

Input tracks 2

Segway parameters • segway --num-labels=25 --segtable=seg_table_min_100. tab --includecoords=encode. Regions. txt. gz --excludecoords=blacklist. female. bed. gz --keep-going --priorstrength=1000 --random-starts=10 --no-posterior --noidentify genomedata. round 6 • segway --num-labels=25 --excludecoords=blacklist. female. bed. gz --mem-usage 8, 10, 12, 14, 15 –d –I input. 0. master –p params. 0. params. 26 –s segway. str --no-train --noposterior genomedata. round 6 3

Segtools report http: //www. ebi. ac. uk/~swilder/segway_round 6_gm 12878_kitchensink/ 4

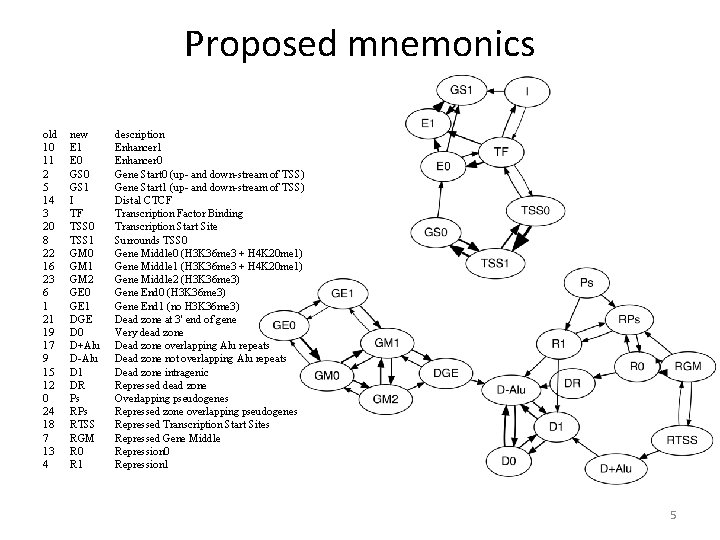
Proposed mnemonics old 10 11 2 5 14 3 20 8 22 16 23 6 1 21 19 17 9 15 12 0 24 18 7 13 4 new E 1 E 0 GS 1 I TF TSS 0 TSS 1 GM 0 GM 1 GM 2 GE 0 GE 1 DGE D 0 D+Alu D-Alu D 1 DR Ps RTSS RGM R 0 R 1 description Enhancer 1 Enhancer 0 Gene Start 0 (up- and down-stream of TSS) Gene Start 1 (up- and down-stream of TSS) Distal CTCF Transcription Factor Binding Transcription Start Site Surrounds TSS 0 Gene Middle 0 (H 3 K 36 me 3 + H 4 K 20 me 1) Gene Middle 1 (H 3 K 36 me 3 + H 4 K 20 me 1) Gene Middle 2 (H 3 K 36 me 3) Gene End 0 (H 3 K 36 me 3) Gene End 1 (no H 3 K 36 me 3) Dead zone at 3' end of gene Very dead zone Dead zone overlapping Alu repeats Dead zone not overlapping Alu repeats Dead zone intragenic Repressed dead zone Overlapping pseudogenes Repressed zone overlapping pseudogenes Repressed Transcription Start Sites Repressed Gene Middle Repression 0 Repression 1 5

K 562 segmentation • Segway short-range K 562 kitchensink (Orion Buske) • Segmentation • Segtools report • Segmentation lift. Over from hg 18 to hg 19 6

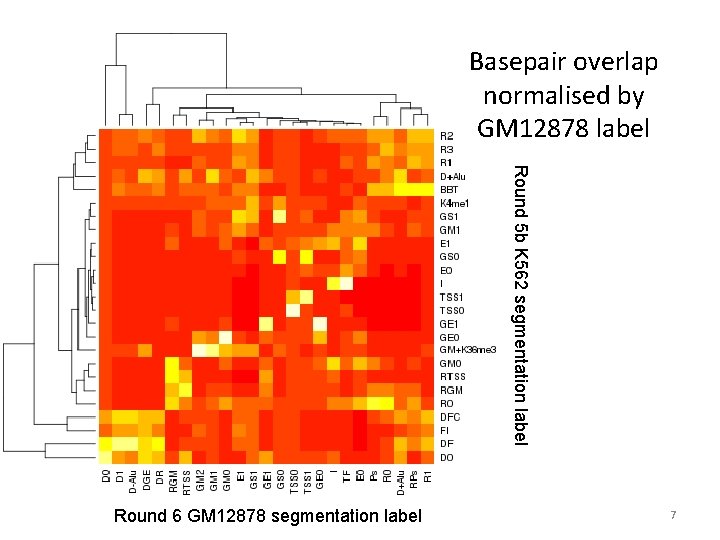
Basepair overlap normalised by GM 12878 label Round 5 b K 562 segmentation label Round 6 GM 12878 segmentation label 7

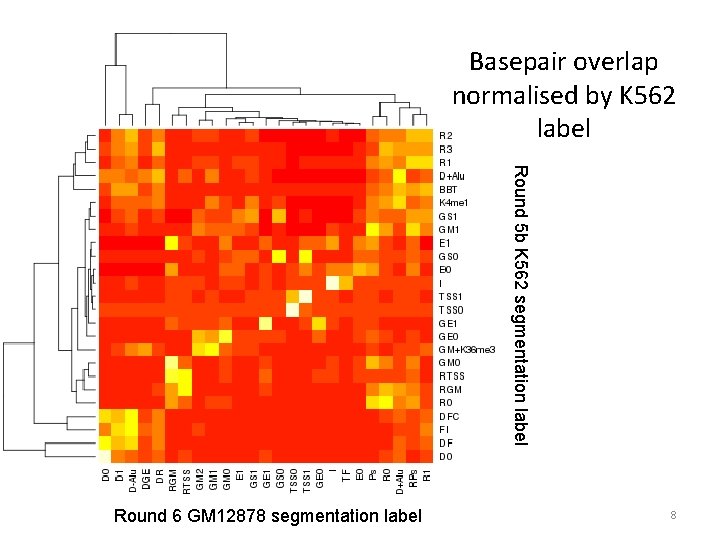
Basepair overlap normalised by K 562 label Round 5 b K 562 segmentation label Round 6 GM 12878 segmentation label 8

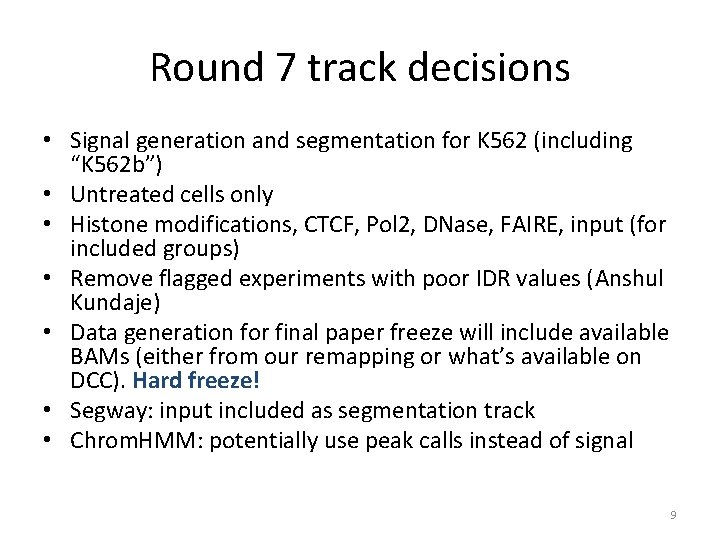
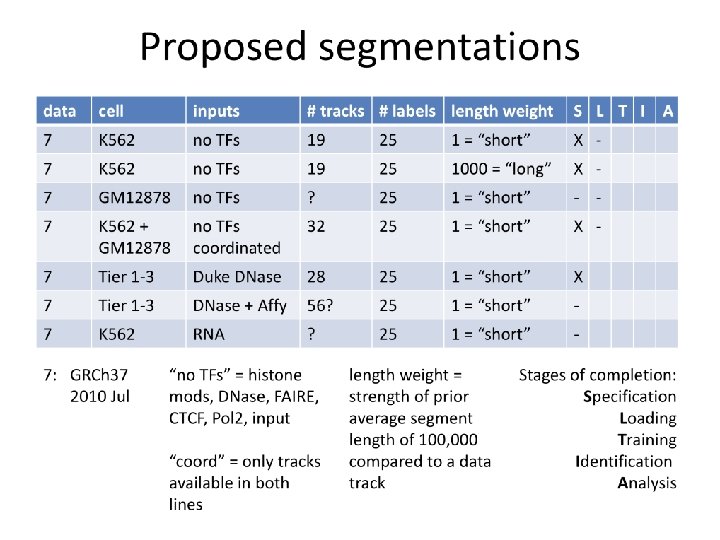
Round 7 track decisions • Signal generation and segmentation for K 562 (including “K 562 b”) • Untreated cells only • Histone modifications, CTCF, Pol 2, DNase, FAIRE, input (for included groups) • Remove flagged experiments with poor IDR values (Anshul Kundaje) • Data generation for final paper freeze will include available BAMs (either from our remapping or what’s available on DCC). Hard freeze! • Segway: input included as segmentation track • Chrom. HMM: potentially use peak calls instead of signal 9

10

Future plans • All data – alignments, signal tracks, peaks, blacklists – stored at EBI • Aspera server running • Segway, Wiggler, Segtools run at EBI • Run “matched” segmentations for Tier 1 and Tier 2 cell lines 11

Acknowledgements • • Ewan Birney Ian Dunham Michael Hoffman Orion Buske 12