hg 19 GRCh 37 vs hg 38 GRCh











- Slides: 29

hg 19 (GRCh 37) vs. hg 38 (GRCh 38) Human Genome Reference Comparison Zuotian Tatum Department of Human Genetics Leiden University Medical Center

Timeline GRCh 37: GRCh 38: First release: Feb 27, 2009 Latest patch: Jun 28, 2013 (p 13) Dec 24, 2013 Latest patch: Oct 14, 2014 (p 1) http: //www. ncbi. nlm. nih. gov/projects/genome/assembly/grc/human/data/

Content GRCh 37. p 13: GRCh 38. p 2: Total bases: N 50: 3. 23 Billion 2. 99 Billion (without N) 46 Million 3. 21 Billion 3. 05 Billion (without N) 67 Million Number of alternative loci: Number of alternative loci : Non-nuclear genome: 9 No 261 Yes http: //www. ncbi. nlm. nih. gov/projects/genome/assembly/grc/human/data/

UCSC tracks for GRCh 38 UCSC Ref. Seq available since April 2014. Ensembl regulatory build available since September 2014. db. SNP 141 available since October 2014. ENCODE and FANTOM 5 track hubs are still not available (Nov 2014).

New in GRCh 38 release Three new sequence files, in addition to the standard assembly files: - GCA_000001405. 15_GRCh 38_top-level. fna. gz - GCA_000001405. 15_GRCh 38_no_alt_analysis_set. fna. gz - GCA_000001405. 15_GRCh 38_full_analysis_set. fna. gz The analysis set files are created to avoid false mapping in NGS alignment pipelines.

GCA_000001405. 15_GRCh 38_top-level. fna. gz All the top-level objects in the full-assembly Chromosomes unlocalized scaffolds unplaced scaffolds alternate locus scaffolds mitochondrial genome The sequence identifiers are International Sequence Database Collaboration (INSDC) accession. versions and the definition lines are Gen. Bank style. No sequences have been hard-masked.

GCA_000001405. 15_GRCh 38_no_alt_analysis_set. fna. gz Chromosomes from the GRCh 38 Primary Assembly unit. Note: the two PAR regions on chr. Y have been hard-masked with Ns. The chromosome Y sequence provided therefore has the same coordinatesas the Gen. Bank sequence but it is not identical to the Gen. Bank sequence. Similarly, duplicate copies of centromeric arrays and WGS on chromosomes 5, 14, 19, 21 & 22 have been hard-masked with Ns. Mitochondrial genome from the GRCh 38 non-nuclear assembly unit. Unlocalized scaffolds from the GRCh 38 Primary Assembly unit. Unplaced scaffolds from the GRCh 38 Primary Assembly unit. Epstein-Barr virus (EBV) sequence Note: The EBV sequence is not part of the genome assembly but isincluded in the analysis set as a sink for alignment of reads that are often present in sequencing samples.

GCA_000001405. 15_GRCh 38_full_analysis_set. fna. gz = GCA_000001405. 15_GRCh 38_no_alt_analysis_set. fna. gz + alt-scaffolds from the GRCh 38 ALT_REF_LOCI_* assembly units

Alt-loci add complexity to RNASeq quantification

Ideogram of GRCh 38. p 2

RNASeq quantification - Fragments (reads) per million per killobase (FPKM/RPKM) values to quantify gene expression - Unique mapping only Analysis tools do not distinguish allelic duplication from paralogous duplication - Non overlapping gene regions

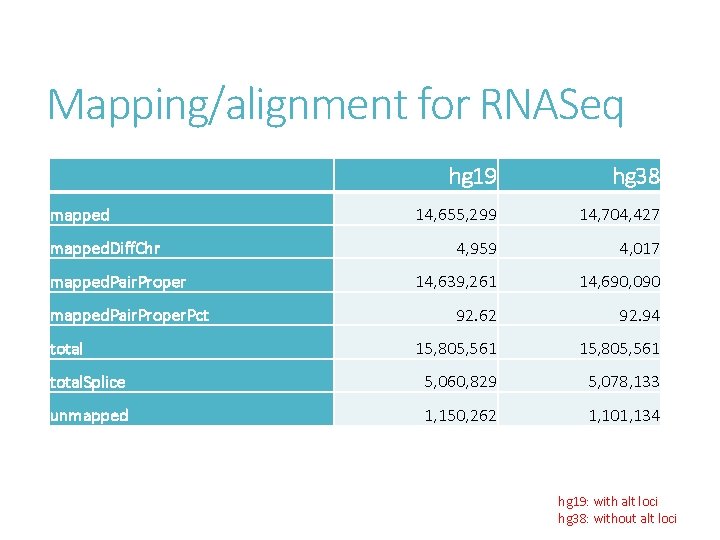
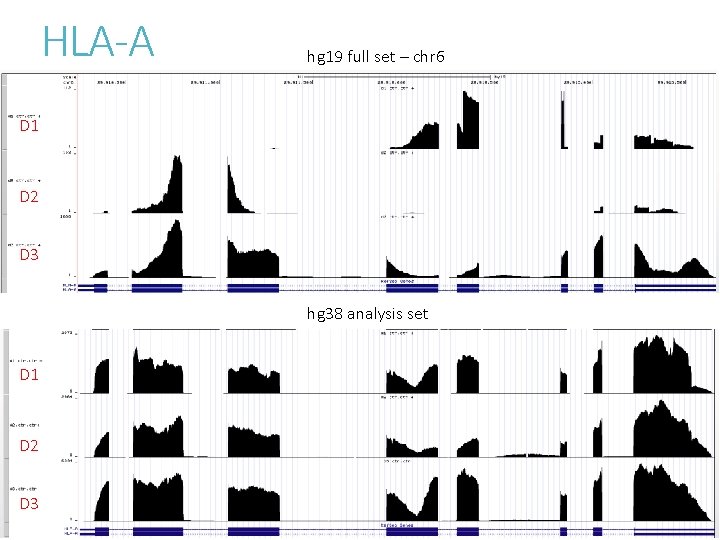
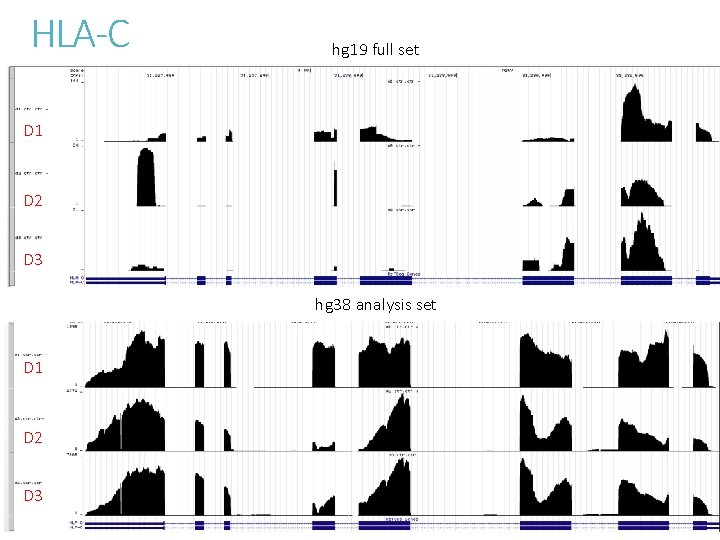
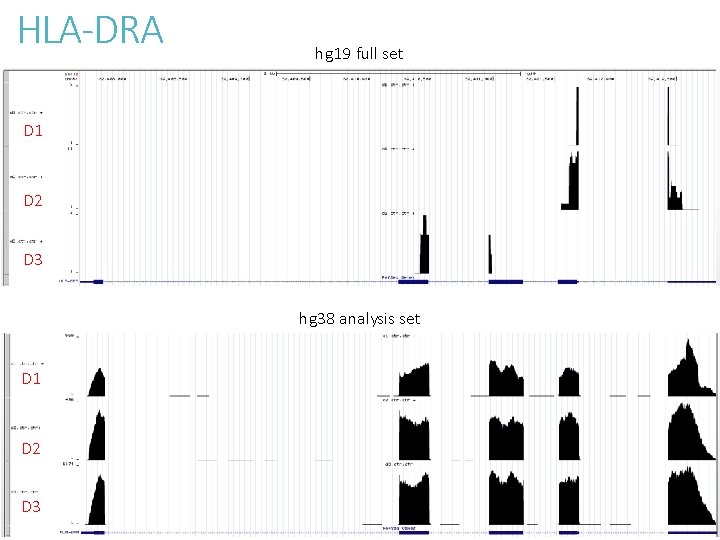
To understand the effect of altloci on RNASeq quantification Compare alignment of chromosome 6 MHC region between - hg 19 full set with 7 alt-loci - hg 38 analysis set without alt-loci Sequence content are largely unchanged between hg 19 and hg 38.

Mapping/alignment for RNASeq hg 19 hg 38 14, 655, 299 14, 704, 427 4, 959 4, 017 14, 639, 261 14, 690, 090 92. 62 92. 94 15, 805, 561 total. Splice 5, 060, 829 5, 078, 133 unmapped 1, 150, 262 1, 101, 134 mapped. Diff. Chr mapped. Pair. Proper. Pct total hg 19: with alt loci hg 38: without alt loci

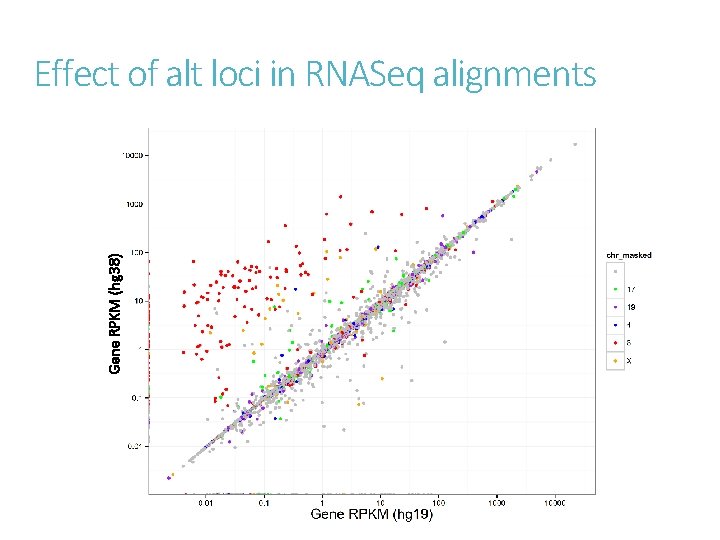
Gene RPKM (hg 38) Effect of alt loci in RNASeq alignments

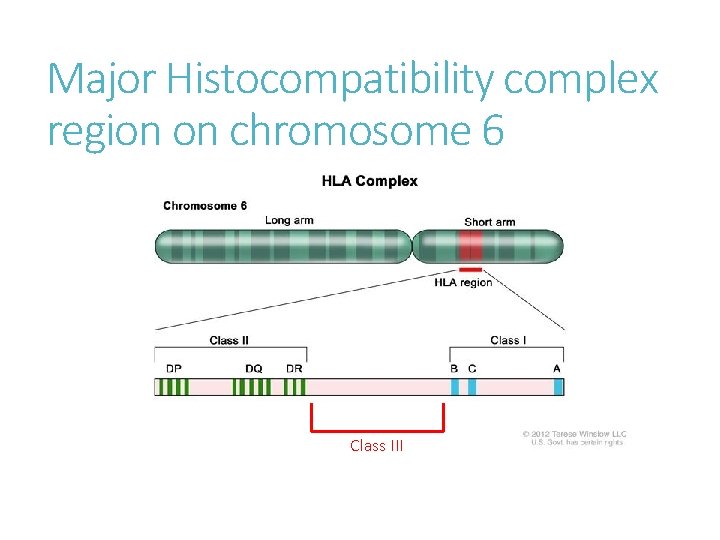
Major Histocompatibility complex region on chromosome 6

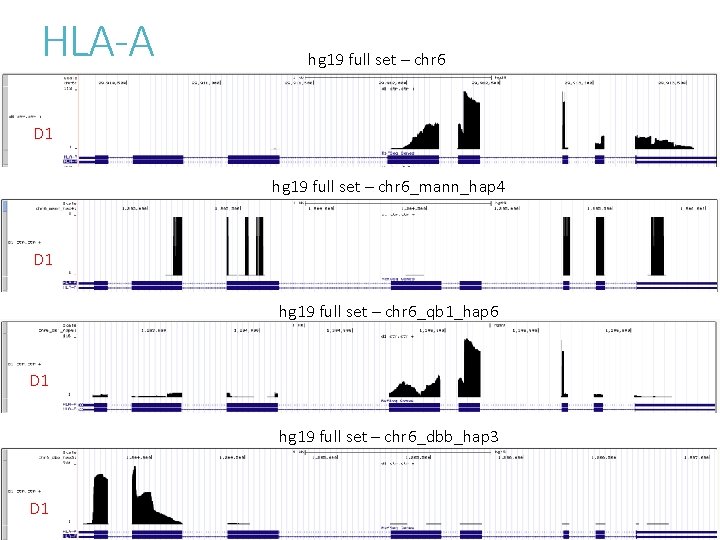
HLA-A hg 19 full set – chr 6 D 1 hg 19 full set – chr 6_mann_hap 4 D 1 hg 19 full set – chr 6_qb 1_hap 6 D 1 hg 19 full set – chr 6_dbb_hap 3 D 1

HLA-A hg 19 full set – chr 6 D 1 D 2 D 3 hg 38 analysis set D 1 D 2 D 3

HLA-C hg 19 full set D 1 D 2 D 3 hg 38 analysis set D 1 D 2 D 3

HLA-DRA hg 19 full set D 1 D 2 D 3 hg 38 analysis set D 1 D 2 D 3

Major Histocompatibility complex region on chromosome 6 Class III

MHC Class III 700 kb stretch, 60 genes. The most gene-dense region of the human genome > 14% coding ~ 72% transcribed Highly conserved Only a free have clearly defined and proven function

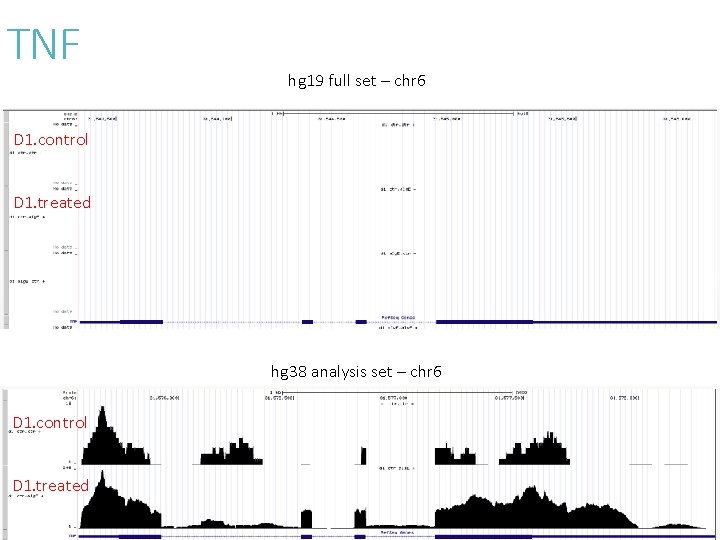
TNF hg 19 full set – chr 6 D 1. control D 1. treated hg 38 analysis set – chr 6 D 1. control D 1. treated

Highly variant immune regions retiled

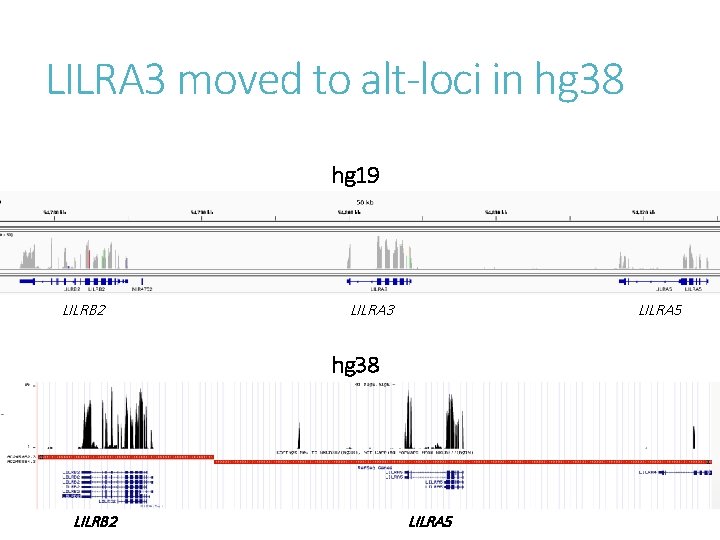
LILRA 3 moved to alt-loci in hg 38 hg 19 LILRB 2 LILRA 3 LILRA 5 hg 38 LILRB 2 LILRA 5

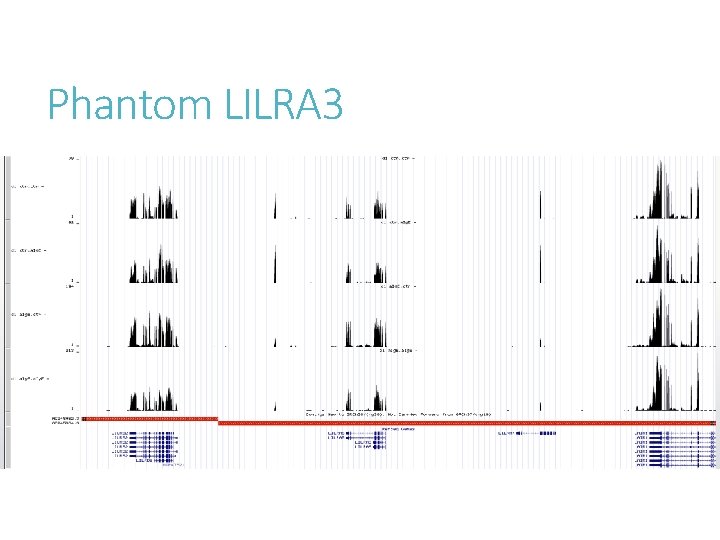
Phantom LILRA 3

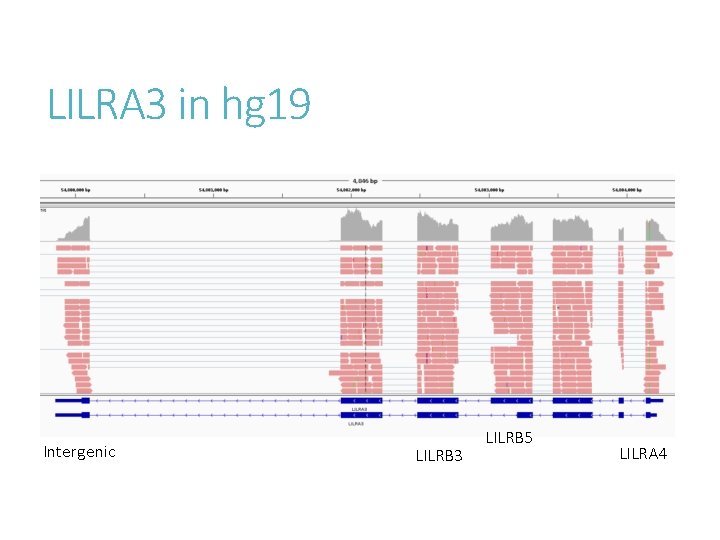
LILRA 3 in hg 19 Intergenic LILRB 3 LILRB 5 LILRA 4

Need more comprehensive approach to genome variation. Assembly model is neither haploid nor diploid Analysis tools penalize reads mapping to > 1 location do not distinguish allelic duplication from paralogous duplication A graph structure is a natural way to represent a populationbased genome assembly

Conclusions RPKM values are highly correlated between hg 19 and hg 38. Analysis set is preferred for expression analysis. Additional analysis may be performed to use the alt-loci separately. Annotations for hg 38 is still lacking and need contribution from the community. Improve modeling of genome variability in population.

Questions?