Genome Annotation of LTR Retrotransposons in Trifolium Repens



- Slides: 8

Genome Annotation of LTR Retrotransposons in Trifolium Repens By: Zain Anwar, Ethan Holleman, Abdullah Mazher, Rohan Rajagopal, and Henry Wittich Under the mentorship of Dr. Howard Laten

Background • White Clover (Trifolium Repens) • Forage crop, native to Europe and Central Asia • Interested in genetic basis of crop domestication • Recent draft assembly published in Gen. Bank • Large influx of sequence data isn’t immediately useful we computationally annotate for LTR retrotransposons

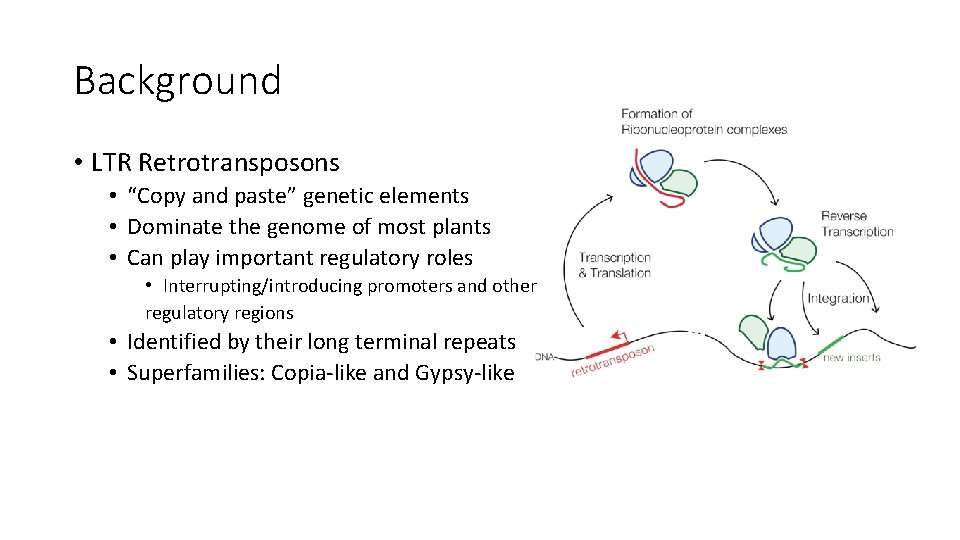
Background • LTR Retrotransposons • “Copy and paste” genetic elements • Dominate the genome of most plants • Can play important regulatory roles • Interrupting/introducing promoters and other regulatory regions • Identified by their long terminal repeats • Superfamilies: Copia-like and Gypsy-like

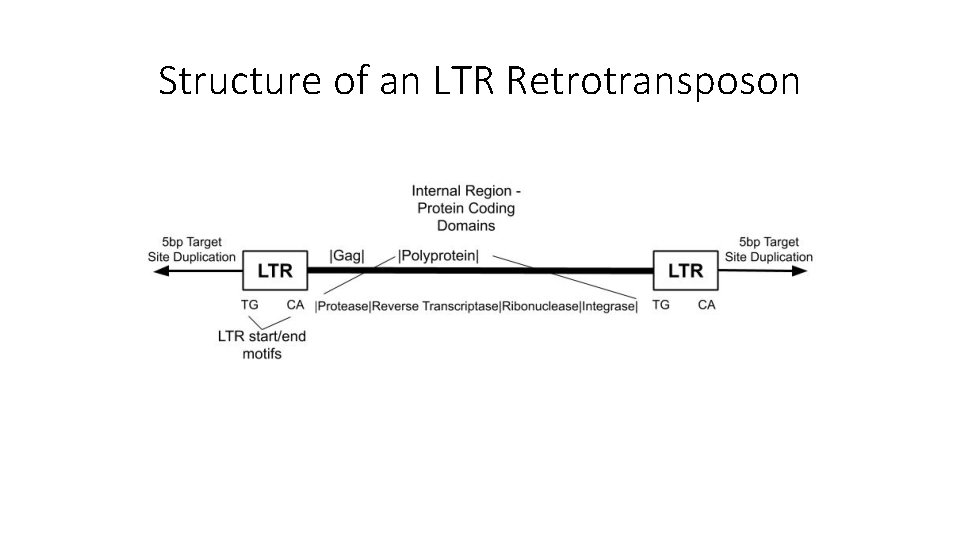
Structure of an LTR Retrotransposon

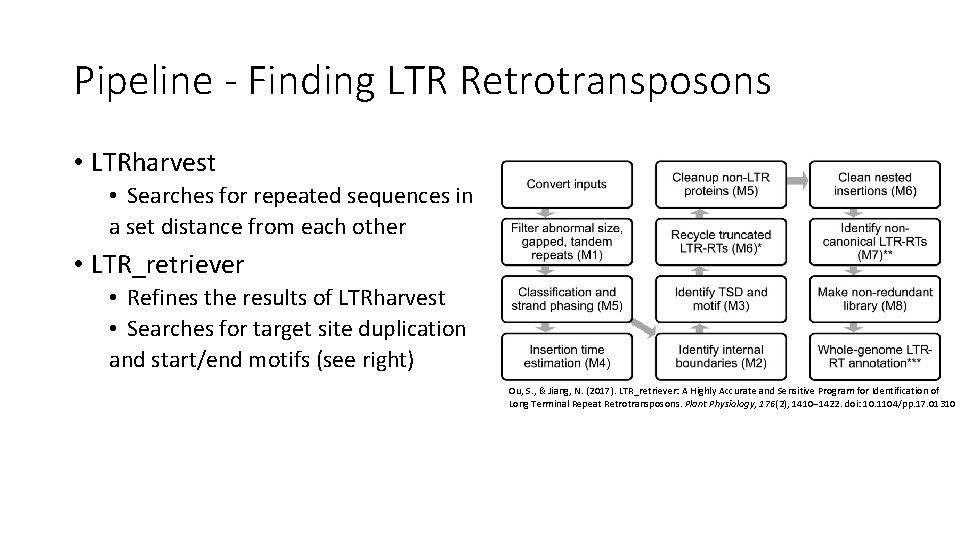
Pipeline - Finding LTR Retrotransposons • LTRharvest • Searches for repeated sequences in a set distance from each other • LTR_retriever • Refines the results of LTRharvest • Searches for target site duplication and start/end motifs (see right) Ou, S. , & Jiang, N. (2017). LTR_retriever: A Highly Accurate and Sensitive Program for Identification of Long Terminal Repeat Retrotransposons. Plant Physiology, 176(2), 1410– 1422. doi: 10. 1104/pp. 17. 01310

Pipeline - Annotating LTR Retrotransposons • Local t. BLASTx against Pfam database • Convert DNA sequence of each LTR retrotransposon into amino acids • Search for protein domains in all 6 reading frames • Compare locations of protein domains in an element • Gypsy elements: protease, reverse transcriptase, ribonuclease, integrase • Copia elements: protease, integrase, reverse transcriptase, ribonuclease

Results So Far • Before filtering out false positives with LTR_retriever, LTRharvest found 2, 730 LTR retrotransposons • This number seemed low; given their abundance in other related plants, it’s expected that there are more in the Trifolium repens genome • After extensive diagnostic tests, it was determined that this low count is a product of the draft assembly being largely incomplete • LTR retrotransposons cluster together and nest within each other, making these highly repetitive regions the most difficult to assemble • Thus far, no annotation has been conducted

Next Steps • Finish designing the annotation portion of the pipeline, including a Python script to run the BLAST searches and classify each element • Run the entire pipeline on the draft assembly to generate test data and tweak pipeline parameters to enhance results • Run the whole pipeline on the complete assembly once it is finished