EVOLUTION OF EUKARYOTIC GENOMES GENE 342 Lecture 13


- Slides: 21

EVOLUTION OF EUKARYOTIC GENOMES GENE 342 Lecture 13 – Comparative genomics

COMPARATIVE GENOMICS • Comparative genomics is a study of the differences and similarities in genome structure and organization in different species • Comparative genomics seek to answer the following questions: 1. How are the differences between humans and other organisms reflected in our genome? and 2. How similar are the number of proteins in humans, fruit flies, worms, plants, yeast and bacteria? • There are two drivers of for comparative genomics 1. Desire to have much more detailed understanding of the process of evolution at the gross level (origin of major classes of organisms) at local level (what make relative species unique) 2. Need to translate DNA sequence data into proteins of known function • The assumption made here is that the DNA sequences encoding important cellular functions are mostly likely to be conserved between species that noncoding sequences

DEFINITIONS Conserved Derived from a common ancestor and retained in contemporary species Evolutionary drift Accumulation of sequences that have little or no impact on fitness Homologues Features which are similar because of being ancestrally related Negative selection Removal of deleterious mutations from a population

DEFINITIONS Phylogenetic distance Measure of the degree of separation between organisms (or their genomes) Can be expressed as accumulated sequence change, number of years, number of generations Positive selection Retention of beneficial mutations Aka Darwinian selection Synteny The property of being on the same chromosome Conserved synteny – genes on the same chromosome in related species (homology blocks)

COMPARATIVE GENOMICS CONT… Orthologues, Paralogues and Gene Displacement • In order to compare genome organization in different organisms it is necessary to distinguish between orthologues and paralogues • Orthologues are homologous genes in different organisms that encode proteins with the same function and which evolved by direct vertical descent • Paralogues are homologous genes within an organism encoding proteins with related but non-identical functions • Orthologues evolve simply by the gradual accumulation of mutations, whereas paralogues arise by gene duplication followed by mutation accumulation

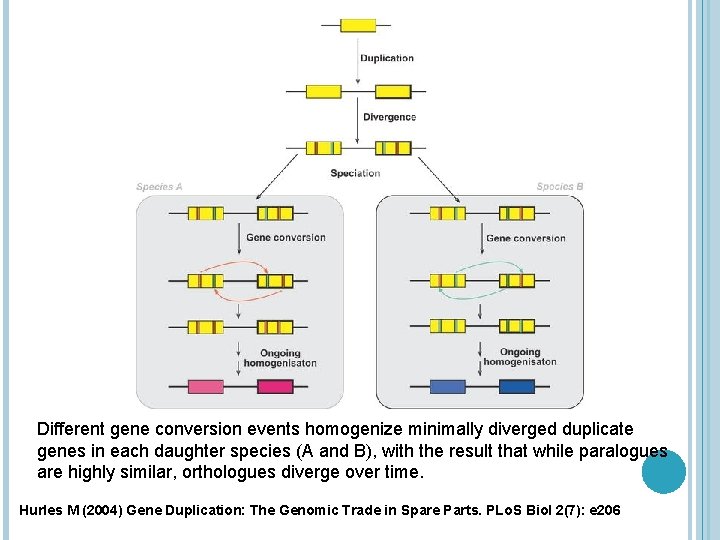
Different gene conversion events homogenize minimally diverged duplicate genes in each daughter species (A and B), with the result that while paralogues are highly similar, orthologues diverge over time. Hurles M (2004) Gene Duplication: The Genomic Trade in Spare Parts. PLo. S Biol 2(7): e 206

COMPARATIVE GENOMICS Orthologues, Paralogues and Gene Displacement • In order to compare genome organization in different organisms it is necessary to distinguish between orthologues and paralogues • Orthologues are homologous genes in different organisms that encode proteins with the same function and which evolved by direct vertical descent • Paralogues are homologous genes within an organism encoding proteins with related but non-identical functions • Orthologues evolve simply by the gradual accumulation of mutations, whereas paralogues arise by gene duplication followed by mutation accumulation • There are many biochemical activities that are common to most or all living organisms, e. g. citric acid cycle, generation of ATP, synthesis of nucleotides, DNA replication etc. • It might be thought that in each case the key proteins would be orthologues. However, there is an increasing evidence that functional equivalence of proteins requires neither sequence similarity nor common three-dimensional folds • Such non-orthologues gene displacement may be rather common, e. g. analysis of the citric acid cycle of enzymes in bacteria and archaea indicate that for at lease 25% of the E. coli enzymes a displacement can be found in at least one other species

COMPARATIVE GENOMICS CONT… Comparative genomics in Eukaryotes Minimal Eukaryotic Genomes • In determining the minimal genome we are seeking to answer number of different questions: 1. What are the fundamental genetic differences between a eukaryotic and prokaryotic cell? 2. What additional genetic information does it require for multicellular coordination? 3. In animals, what are the minimum sizes for a vertebrate genome and a mammalian genome? 4. What is the minimum size of genome for a flowering plant? • Given that many eukaryotic genomes contain large amounts of non-coding DNA, these question has to be answered by considering both genome size and the number of proteins that are encoded: • Only limited number of genomes from microbial eukaryotes that have been sequenced, which makes it difficult to specify what constitute the minimal genome size for a free-living eukaryote

COMPARATIVE GENOMICS CONT… Comparative genomics in Eukaryotes Minimal Eukaryotic Genomes • However, a eukaryotic parasite, Encephalitozoon cuiculi, has a genome of only 2. 9 Mb and that of a close relative, Encephalitozoon instestinalis, ~ 2. 3 Mb • These genomes contain very little repetitive DNA other than r. DNA and probably contain less than 2000 genes. This is still 7 -8 times greater than small prokaryotic genome and it is hard to predict how many of these genes will be involved in mitosis and related events • Nevertheless, the change from a prokaryote to a eukaryote does not require a major increase in genome size • Of the multicellular organisms whose genomes have been sequenced, Arabidopsis, Caenorhabditis and Drosophila encode approximately similar numbers of proteins (11000 -18000) • Among vertebrates, Japanese puffer fish (Fugu rubripes) has the smallest genome identified to date but has a similar gene repertoire to other vertebrates such as humans • Whereas about 35000 genes are spread over 3000 Mb of DNA in human genome, these same genes are restricted to just 400 Mb

Arabidopsis thaliana Caenorhabditis elegans Human beings

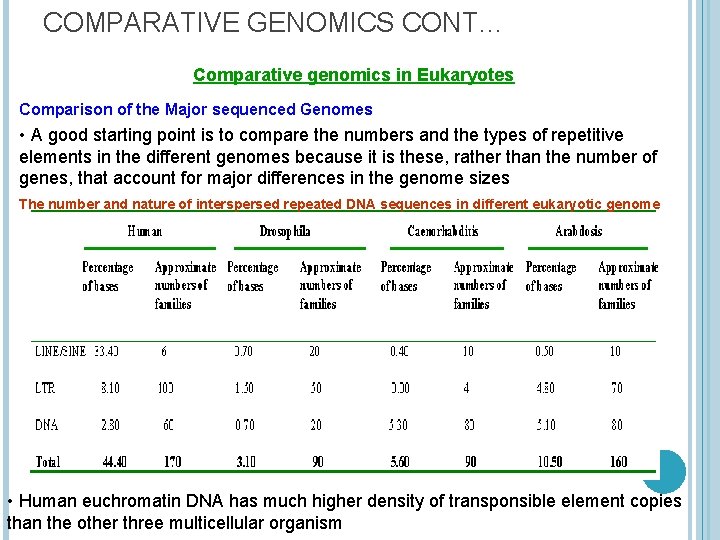
COMPARATIVE GENOMICS CONT… Comparative genomics in Eukaryotes Comparison of the Major sequenced Genomes • A good starting point is to compare the numbers and the types of repetitive elements in the different genomes because it is these, rather than the number of genes, that account for major differences in the genome sizes The number and nature of interspersed repeated DNA sequences in different eukaryotic genome • Human euchromatin DNA has much higher density of transponsible element copies than the other three multicellular organism

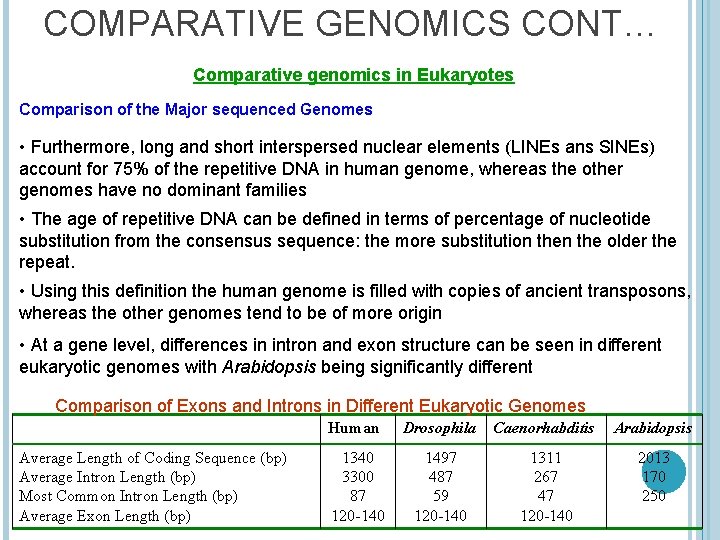
COMPARATIVE GENOMICS CONT… Comparative genomics in Eukaryotes Comparison of the Major sequenced Genomes • Furthermore, long and short interspersed nuclear elements (LINEs ans SINEs) account for 75% of the repetitive DNA in human genome, whereas the other genomes have no dominant families • The age of repetitive DNA can be defined in terms of percentage of nucleotide substitution from the consensus sequence: the more substitution the older the repeat. • Using this definition the human genome is filled with copies of ancient transposons, whereas the other genomes tend to be of more origin • At a gene level, differences in intron and exon structure can be seen in different eukaryotic genomes with Arabidopsis being significantly different Comparison of Exons and Introns in Different Eukaryotic Genomes Average Length of Coding Sequence (bp) Average Intron Length (bp) Most Common Intron Length (bp) Average Exon Length (bp) Human Drosophila Caenorhabditis Arabidopsis 1340 3300 87 120 -140 1497 487 59 120 -140 1311 267 47 120 -140 2013 170 250

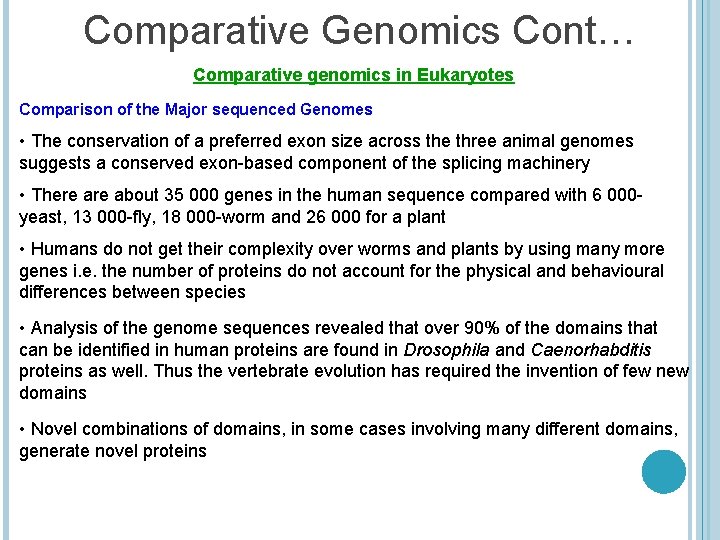
Comparative Genomics Cont… Comparative genomics in Eukaryotes Comparison of the Major sequenced Genomes • The conservation of a preferred exon size across the three animal genomes suggests a conserved exon-based component of the splicing machinery • There about 35 000 genes in the human sequence compared with 6 000 yeast, 13 000 -fly, 18 000 -worm and 26 000 for a plant • Humans do not get their complexity over worms and plants by using many more genes i. e. the number of proteins do not account for the physical and behavioural differences between species • Analysis of the genome sequences revealed that over 90% of the domains that can be identified in human proteins are found in Drosophila and Caenorhabditis proteins as well. Thus the vertebrate evolution has required the invention of few new domains • Novel combinations of domains, in some cases involving many different domains, generate novel proteins

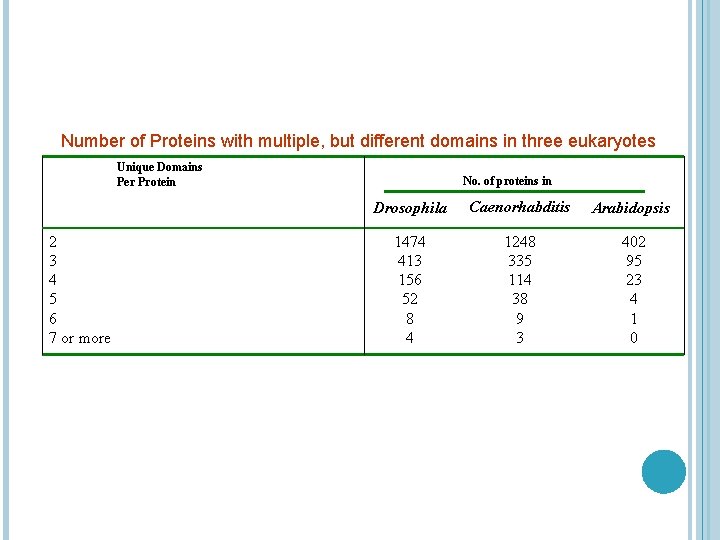
Number of Proteins with multiple, but different domains in three eukaryotes Unique Domains Per Protein 2 3 4 5 6 7 or more No. of proteins in Drosophila Caenorhabditis Arabidopsis 1474 413 156 52 8 4 1248 335 114 38 9 3 402 95 23 4 1 0

Comparative Genomics Cont… Comparative genomics in Eukaryotes Comparison of the Major sequenced Genomes • A high percentage of proteins encoded by any one of the sequenced genomes have orthologues in the other sequenced genomes. For example, 60% of the predicted human proteins have sequence similarities to yeast, fly, nematode or plant proteins and 61% of fly proteins 43% of nematode proteins and 46% of yeast proteins have human counterparts • The functions that differ significantly between humans and the other sequenced genomes are those involved in acquired immunity, neural development, intercellular and intracellular signaling, haemostasis and apoptosis

COMPARATIVE GENOMICS • A complete genome sequence of an organism can be considered to be the ultimate genetic map, in the sense that: 1. Heritable characteristics are encoded within DNA 2. Order of all nucleotides along each chromosome is known 3. Most of the genes present in the genome are identified • However, knowledge of the DNA sequences does not tell us directly how this genetic information to the observable traits and behaviors (phenotypes) that we want to understand i. e. it does not tell us the functions of many of these genes and the importance of these genes to the life of the cell • Finding all the functional parts of the genome sequences and using this information to improve the health of individuals and society are the focus of the next phase of the human genome project • Comparative analysis will be a major part of this effort • The major principles of comparative genomics are straightforward. Common features o two organisms will often be encoded within the DNA that is conserved between the species

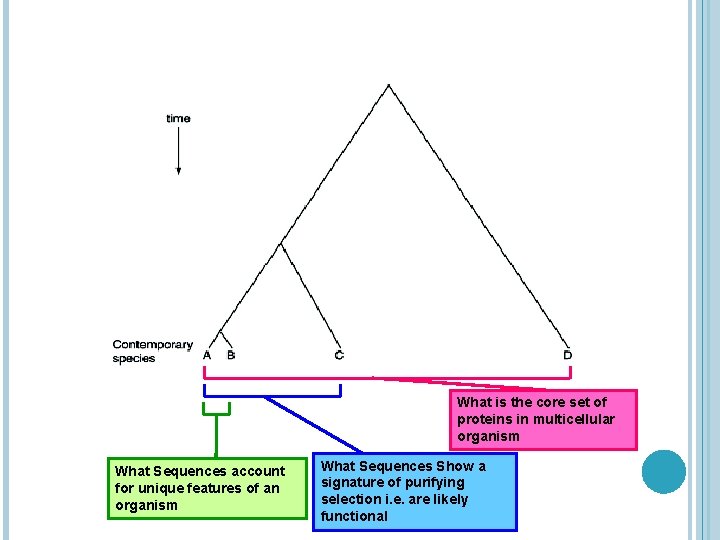
COMPARATIVE GENOMICS CONT… • The DNA sequences encoding the proteins and RNAs responsible for functions that were conserved from the last common ancestor should be preserved in contemporary genome sequences • Likewise, the DNA sequences controlling the expression of genes that are regulated similarly in in two related species should also be conserved • Conversely, sequences that encode (or control the expression of) proteins and RNAs responsible for differences between species will themselves be divergent • Different questions can be answered by comparing genomes at different phylogenetic distances (see diagram)

What is the core set of proteins in multicellular organism What Sequences account for unique features of an organism What Sequences Show a signature of purifying selection i. e. are likely functional

Comparative Genomics Cont… • The DNA sequences the proteins and RNAs responsible for functions that were conserved from the last common ancestor should be preserved in contemporary genome sequences • Likewise, the DNA sequences controlling the expression of genes that are regulated similarly in in two related species should also be conserved • Conversely, sequences that encode (or control the expression of) proteins and RNAs responsible for differences between species will themselves be divergent • Different questions can be answered by comparing genomes at different phylogenetic distances • Broad insights about types of genes can be gleaned by genomic comparisons at very long phylogenetic distances, e. g. , > 1 billion years since their separation • For example, comparing genomes of yeast, worms and flies reveals that these eukaryotes: 1. Encodes many of the same proteins 2. Non-repetitive protein sets of flies and worms are about the same size, being only twice that of yeast

COMPARATIVE GENOMICS CONT… • Over such very large distances, the order of genes and the sequences regulating their expression are generally not conserved. • At moderate phylogenetic distances (~ 70 -100 million years of divergence) both function and non-functional DNA is found within the conserved region • In these cases, the functional sequences will show a signature of purifying or negative selection = the functional sequences will have changed less that the non-functional or neutral DNA • In addition to discriminate conserved from divergent and functional from nonfunctional DNA, comparative genomics is also contributing to identifying the general functional class of certain DNA segments, such as: 1. Coding exons 2. Non-coding RNA, and 3. Some genes regulatory regions • Examples of analysis at this distance (~ 70 -100 mln yrs) include comparisons among enteric bacteria, several species of yeast, and between mouse and human (see diagrams)

COMPARATIVE GENOMICS CONT… • In contrast, very similar genomes, such as those of humans and chimpanzees (separated by 5 billion years of evolution), are particularly apt for finding the key sequence differences that may account for differences in organisms • These are sequence changes under positive selection = the retention of mutations that benefit an organism; also referred to as Darwinian selection • Comparative Genomics is thus a powerful and burgeoning discipline that becomes more and more informative as genomic sequence data accumulate