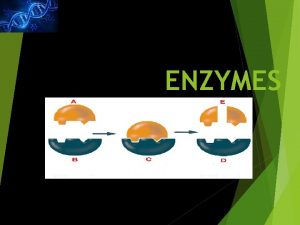
Enzymes A Molecular Perspective Shuchismita Dutta Ph D
- Slides: 18

Enzymes: A Molecular Perspective Shuchismita Dutta, Ph. D.

Learning Objectives • Introduction to Enzymes • Enzyme action • Enzyme regulation Developed as part of the RCSB Collaborative Curriculum Development Program 2016

Learning Objectives • Introduction to Enzymes • Enzyme action • Enzyme regulation Developed as part of the RCSB Collaborative Curriculum Development Program 2016

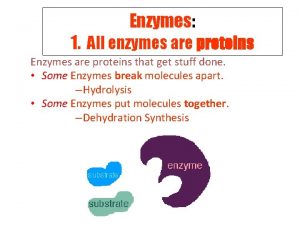
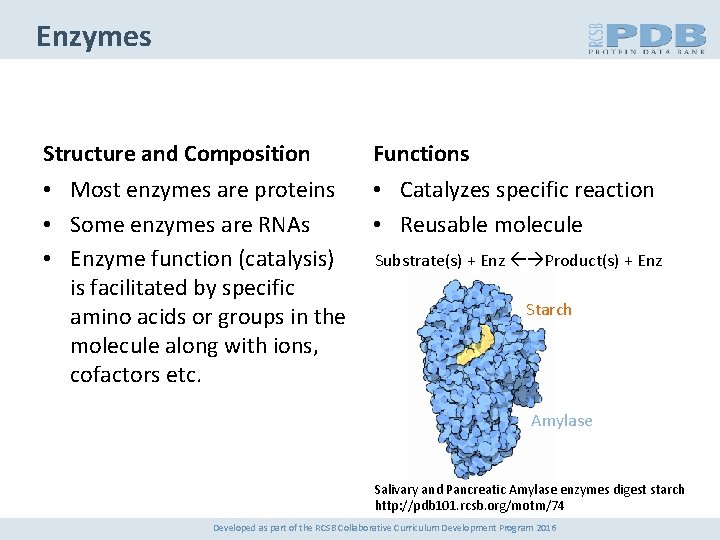
Enzymes Structure and Composition Functions • Most enzymes are proteins • Some enzymes are RNAs • Enzyme function (catalysis) is facilitated by specific amino acids or groups in the molecule along with ions, cofactors etc. • Catalyzes specific reaction • Reusable molecule Substrate(s) + Enz Product(s) + Enz Starch Amylase Salivary and Pancreatic Amylase enzymes digest starch http: //pdb 101. rcsb. org/motm/74 Developed as part of the RCSB Collaborative Curriculum Development Program 2016

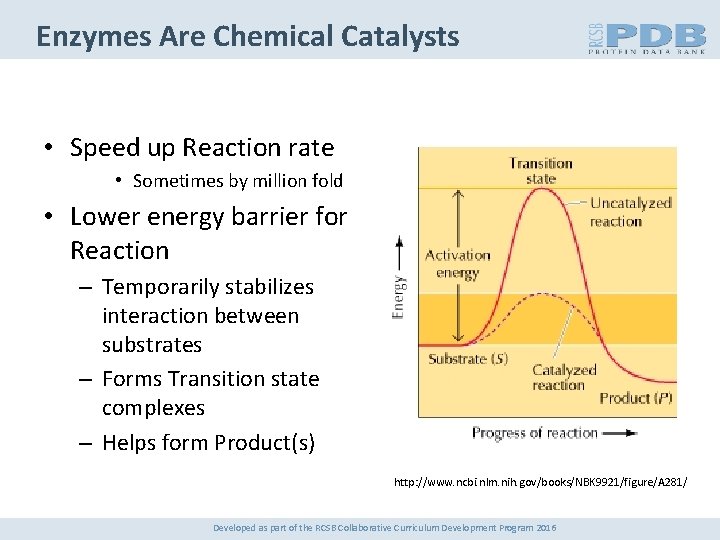
Enzymes Are Chemical Catalysts • Speed up Reaction rate • Sometimes by million fold • Lower energy barrier for Reaction – Temporarily stabilizes interaction between substrates – Forms Transition state complexes – Helps form Product(s) http: //www. ncbi. nlm. nih. gov/books/NBK 9921/figure/A 281/ Developed as part of the RCSB Collaborative Curriculum Development Program 2016

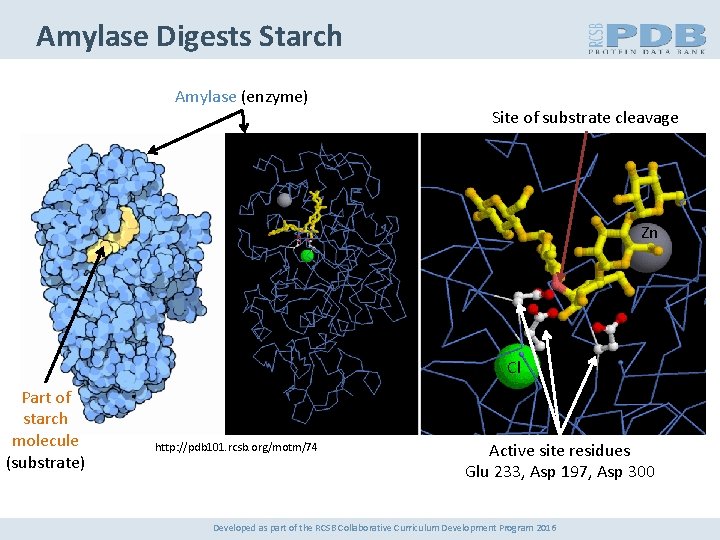
Amylase Digests Starch Amylase (enzyme) Site of substrate cleavage Zn Cl Part of starch molecule (substrate) http: //pdb 101. rcsb. org/motm/74 Active site residues Glu 233, Asp 197, Asp 300 Developed as part of the RCSB Collaborative Curriculum Development Program 2016

Learning Objectives • Introduction to Enzymes • Enzyme action • Enzyme regulation Developed as part of the RCSB Collaborative Curriculum Development Program 2016

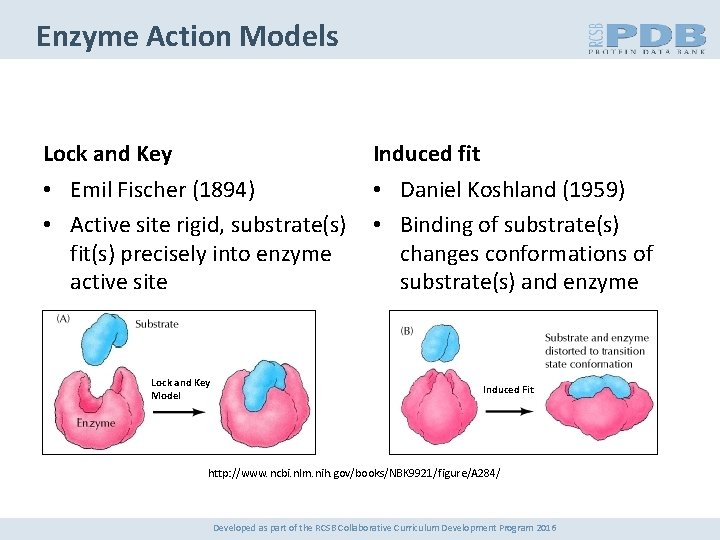
Enzyme Action Models Lock and Key Induced fit • Emil Fischer (1894) • Active site rigid, substrate(s) fit(s) precisely into enzyme active site • Daniel Koshland (1959) • Binding of substrate(s) changes conformations of substrate(s) and enzyme Lock and Key Model Induced Fit http: //www. ncbi. nlm. nih. gov/books/NBK 9921/figure/A 284/ Developed as part of the RCSB Collaborative Curriculum Development Program 2016

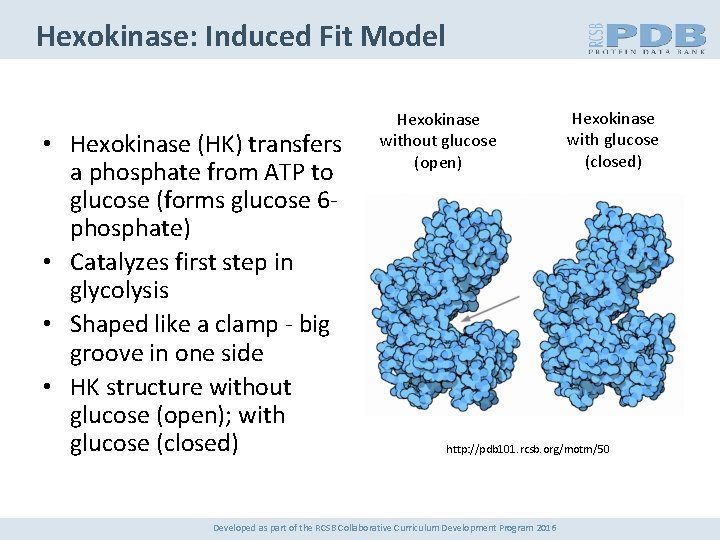
Hexokinase: Induced Fit Model • Hexokinase (HK) transfers a phosphate from ATP to glucose (forms glucose 6 phosphate) • Catalyzes first step in glycolysis • Shaped like a clamp - big groove in one side • HK structure without glucose (open); with glucose (closed) Hexokinase without glucose (open) Hexokinase with glucose (closed) http: //pdb 101. rcsb. org/motm/50 Developed as part of the RCSB Collaborative Curriculum Development Program 2016

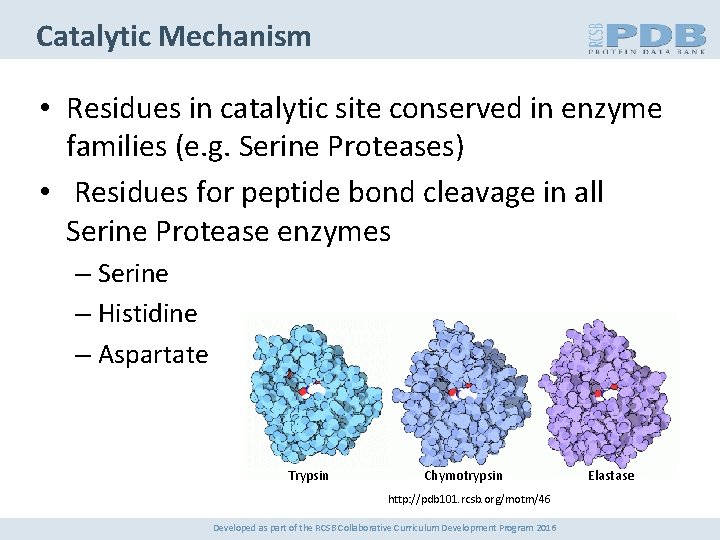
Catalytic Mechanism • Residues in catalytic site conserved in enzyme families (e. g. Serine Proteases) • Residues for peptide bond cleavage in all Serine Protease enzymes – Serine – Histidine – Aspartate Trypsin Chymotrypsin http: //pdb 101. rcsb. org/motm/46 Developed as part of the RCSB Collaborative Curriculum Development Program 2016 Elastase

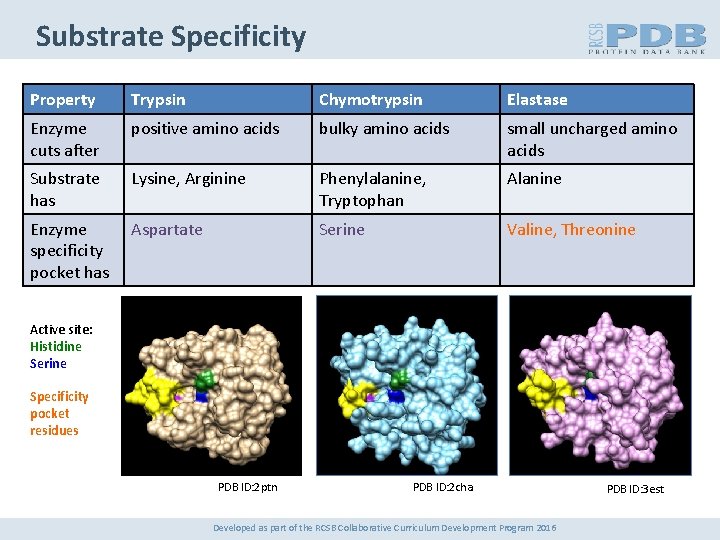
Substrate Specificity Property Trypsin Chymotrypsin Elastase Enzyme cuts after positive amino acids bulky amino acids small uncharged amino acids Substrate has Lysine, Arginine Phenylalanine, Tryptophan Alanine Enzyme specificity pocket has Aspartate Serine Valine, Threonine Active site: Histidine Serine Specificity pocket residues PDB ID: 2 ptn PDB ID: 2 cha Developed as part of the RCSB Collaborative Curriculum Development Program 2016 PDB ID: 3 est

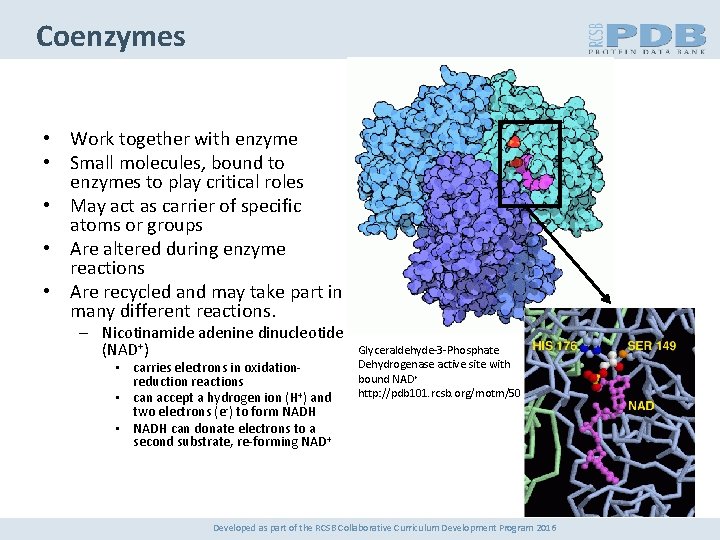
Coenzymes • Work together with enzyme • Small molecules, bound to enzymes to play critical roles • May act as carrier of specific atoms or groups • Are altered during enzyme reactions • Are recycled and may take part in many different reactions. – Nicotinamide adenine dinucleotide (NAD+) • carries electrons in oxidationreduction reactions • can accept a hydrogen ion (H+) and two electrons (e-) to form NADH • NADH can donate electrons to a second substrate, re-forming NAD+ Glyceraldehyde-3 -Phosphate Dehydrogenase active site with bound NAD+ http: //pdb 101. rcsb. org/motm/50 Developed as part of the RCSB Collaborative Curriculum Development Program 2016

Learning Objectives • Introduction to Enzymes • Enzyme action • Enzyme regulation Developed as part of the RCSB Collaborative Curriculum Development Program 2016

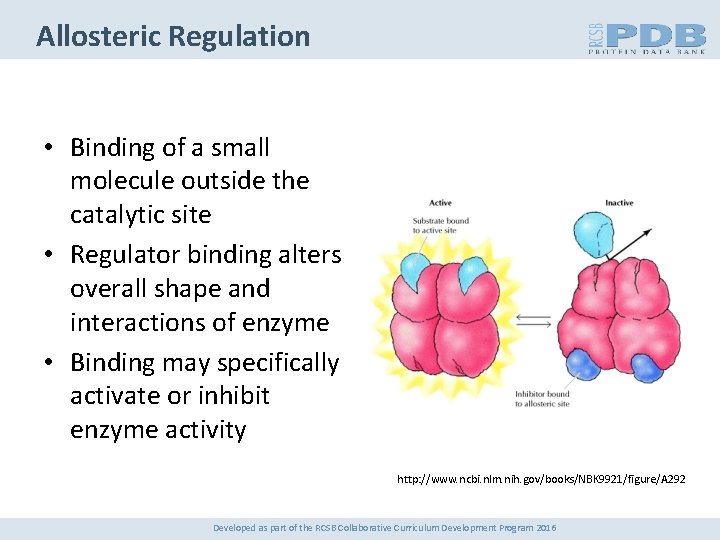
Allosteric Regulation • Binding of a small molecule outside the catalytic site • Regulator binding alters overall shape and interactions of enzyme • Binding may specifically activate or inhibit enzyme activity http: //www. ncbi. nlm. nih. gov/books/NBK 9921/figure/A 292 Developed as part of the RCSB Collaborative Curriculum Development Program 2016

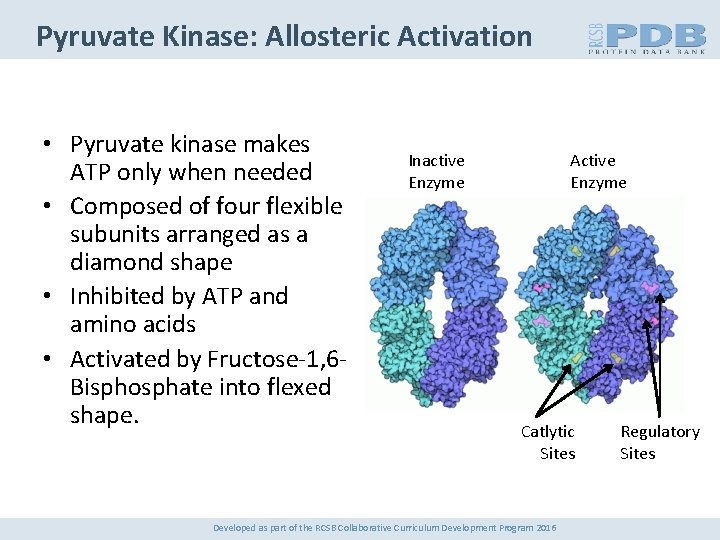
Pyruvate Kinase: Allosteric Activation • Pyruvate kinase makes ATP only when needed • Composed of four flexible subunits arranged as a diamond shape • Inhibited by ATP and amino acids • Activated by Fructose-1, 6 Bisphosphate into flexed shape. Inactive Enzyme Active Enzyme Catlytic Sites Developed as part of the RCSB Collaborative Curriculum Development Program 2016 Regulatory Sites

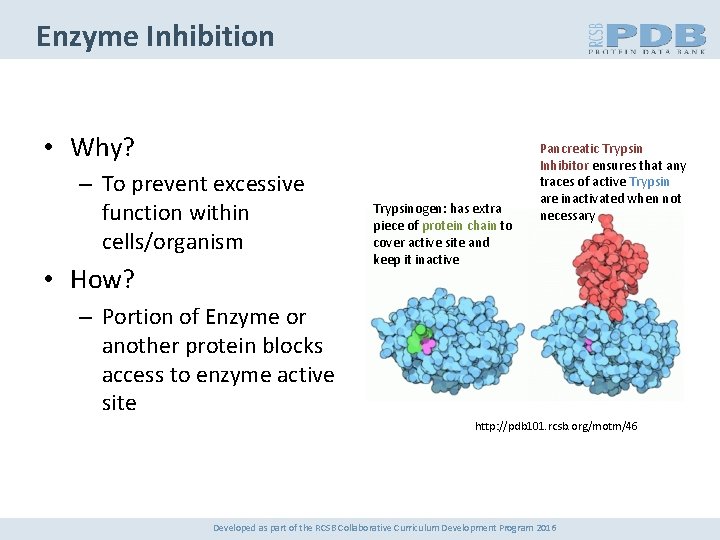
Enzyme Inhibition • Why? – To prevent excessive function within cells/organism • How? Trypsinogen: has extra piece of protein chain to cover active site and keep it inactive Pancreatic Trypsin Inhibitor ensures that any traces of active Trypsin are inactivated when not necessary – Portion of Enzyme or another protein blocks access to enzyme active site http: //pdb 101. rcsb. org/motm/46 Developed as part of the RCSB Collaborative Curriculum Development Program 2016

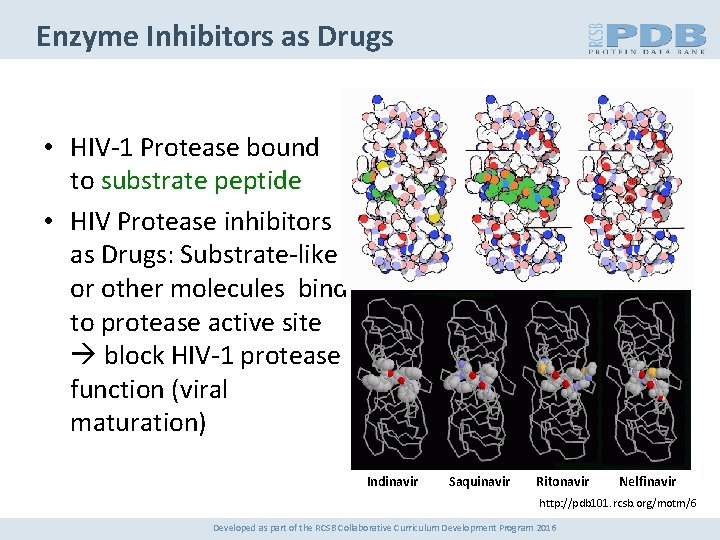
Enzyme Inhibitors as Drugs • HIV-1 Protease bound to substrate peptide • HIV Protease inhibitors as Drugs: Substrate-like or other molecules bind to protease active site block HIV-1 protease function (viral maturation) Indinavir Saquinavir Ritonavir Nelfinavir http: //pdb 101. rcsb. org/motm/6 Developed as part of the RCSB Collaborative Curriculum Development Program 2016

Summary • Introduction to Enzymes – Enzymes and Catalysis • Enzyme action – Enzyme Action models – Catalytic mechanisms, specificity and coenzymes • Enzyme regulation – Allosteric Regulation – Inhibition and Drugs Developed as part of the RCSB Collaborative Curriculum Development Program 2016
 Niloy dutta uconn
Niloy dutta uconn Dipanwita dutta
Dipanwita dutta Chris keane wsu
Chris keane wsu Dr s k dutta
Dr s k dutta Anup dutta hcl
Anup dutta hcl Partha dutta rpi
Partha dutta rpi Ahb hprot
Ahb hprot Giant molecular structure vs simple molecular structure
Giant molecular structure vs simple molecular structure Zinc oxide + nitric acid → zinc nitrate + water
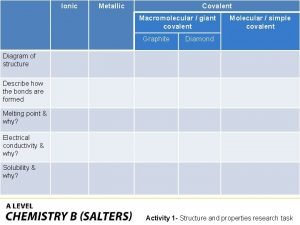
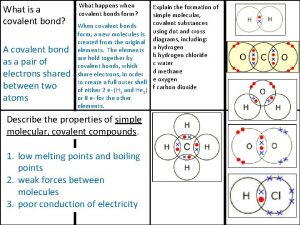
Zinc oxide + nitric acid → zinc nitrate + water Covalent bond melting point
Covalent bond melting point Silo perspective vs business process perspective
Silo perspective vs business process perspective One point perspective house
One point perspective house The principal enzyme involved in dna replication is
The principal enzyme involved in dna replication is 5 enzymes responsible for dna replication
5 enzymes responsible for dna replication Metobolic encephalopathy
Metobolic encephalopathy Steady state biochemistry
Steady state biochemistry Substrate of cellular respiration
Substrate of cellular respiration Elevated liver enzymes causes
Elevated liver enzymes causes Restriction enzymes
Restriction enzymes