Chromosome 2 Doil Choi Sunghwan Jo KOREA Cytological


- Slides: 11

Chromosome 2 Doil Choi, Sunghwan Jo KOREA

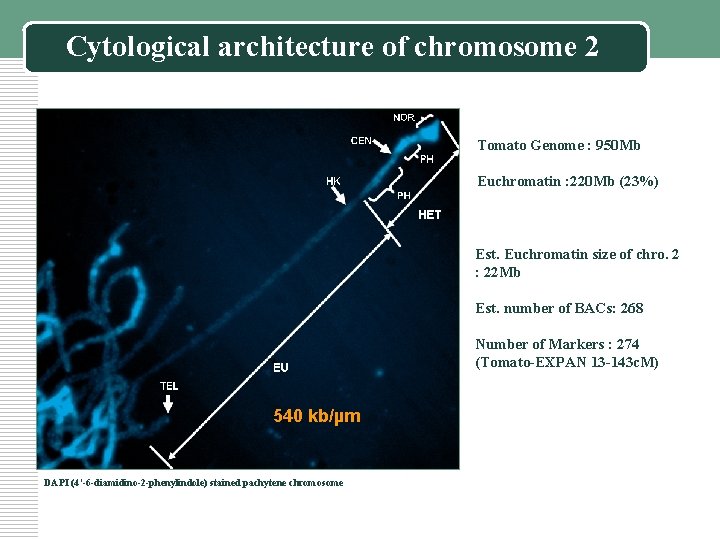
Cytological architecture of chromosome 2 Tomato Genome : 950 Mb Euchromatin : 220 Mb (23%) Est. Euchromatin size of chro. 2 : 22 Mb Est. number of BACs: 268 Number of Markers : 274 (Tomato-EXPAN 13 -143 c. M) 540 kb/µm DAPI (4’-6 -diamidino-2 -phenylindole) stained pachytene chromosome

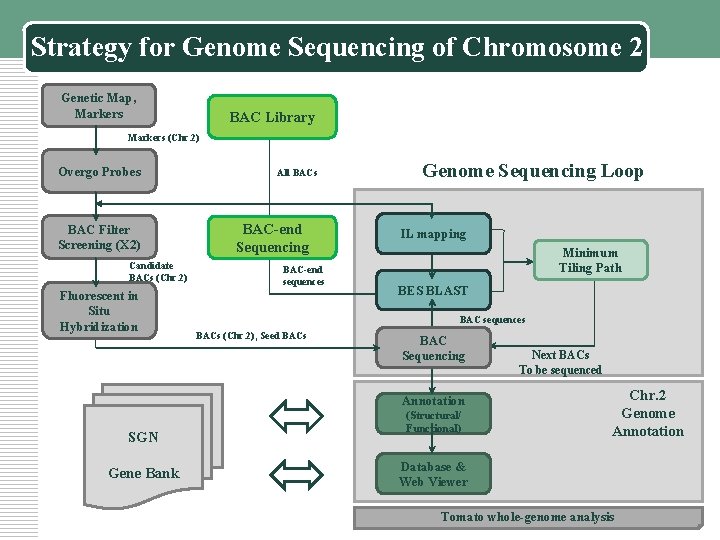
Strategy for Genome Sequencing of Chromosome 2 Genetic Map, Markers BAC Library Markers (Chr. 2) Overgo Probes BAC Filter Screening (X 2) Candidate BACs (Chr. 2) Fluorescent in Situ Hybridization All BACs BAC-end Sequencing BAC-end sequences Genome Sequencing Loop IL mapping Minimum Tiling Path BES BLAST BAC sequences BACs (Chr. 2), Seed BACs BAC Sequencing Annotation SGN Gene Bank (Structural/ Functional) Next BACs To be sequenced Chr. 2 Genome Annotation Database & Web Viewer Tomato whole-genome analysis

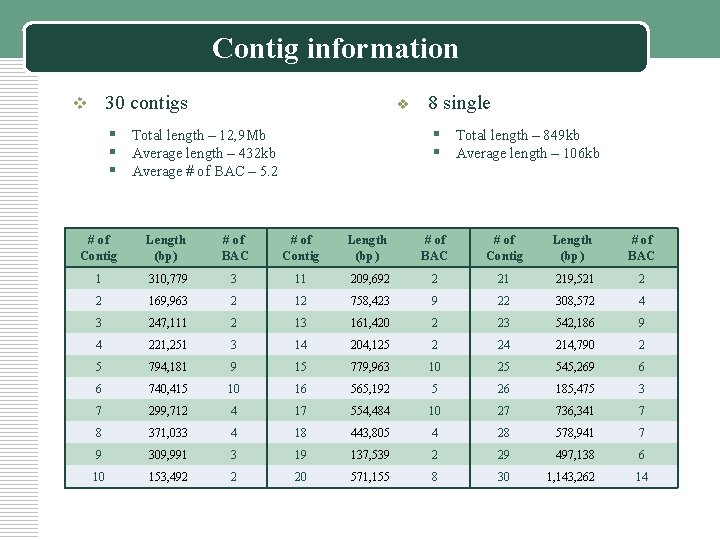
Contig information v 30 contigs v § Total length – 12, 9 Mb § Average length – 432 kb § Average # of BAC – 5. 2 8 single § Total length – 849 kb § Average length – 106 kb # of Contig Length (bp) # of BAC 1 310, 779 3 11 209, 692 2 21 219, 521 2 2 169, 963 2 12 758, 423 9 22 308, 572 4 3 247, 111 2 13 161, 420 2 23 542, 186 9 4 221, 251 3 14 204, 125 2 24 214, 790 2 5 794, 181 9 15 779, 963 10 25 545, 269 6 6 740, 415 10 16 565, 192 5 26 185, 475 3 7 299, 712 4 17 554, 484 10 27 736, 341 7 8 371, 033 4 18 443, 805 4 28 578, 941 7 9 309, 991 3 19 137, 539 2 29 497, 138 6 10 153, 492 2 20 571, 155 8 30 1, 143, 262 14

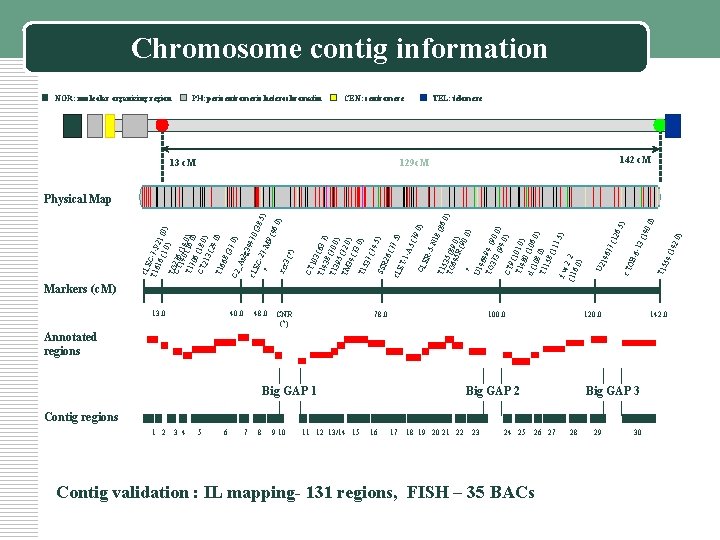
1 2 13. 0 40. 0 3 4 5 6 48. 0 7 8 9 10 CNR (? ) 78. 0 11 12 13/14 15 16 100. 0 Big GAP 1 Big GAP 2 17 18 19 20 21 22 23 24 25 26 27 Contig validation : IL mapping- 131 regions, FISH – 35 BACs 28 29 120. 0 Annotated regions Big GAP 3 Contig regions 30 ) 42. 0 4 (1 T 15 5 0) 40. 3 (1 5) 129 c. M B-6 -1 26. 13 c. M c. TO CEN: centromere 677 (1 PH: pericentromeric heterochromatin U 2 14 NOR: nucleolar organizing region CT 103 T 14 (63. 7 ) 38 T 13 (70. 0 ) 95 TM (72. 0 ) 34 (73. 0) T 15 37 (74. 5) SSR 26 (77. 5) c. LE T-1 -A 5 (79. 0) CL ER -5 -N 18 (86 T 15. 0) TG 35 (8 645 9. 0) R( 90. 0) ? U 1 464 TG 94 ( 90 373 (94. 0) CT 9 (1 0 1 T 14. 80 0) d (1 (106. 0) 08 T 11. 0) 58 (11 1. 5 ) f. w. 2. 2 (11 6. 0 ) c. LE T 16 C-7 -P 2 16 (1. 0 1 (0) ) TG CT 276 140 (14 T 17 (16. . 00) ) 06 CT (18. 0 213 ) (24. 0) T 16 68 (37. 0) C 2_ At 2 g 34 470 c. LE (38 C-2. 5) 7 M ? 9 (4 6. 0 ) rcr 3 (? ) Chromosome contig information TEL: telomere 142 c. M Physical Map Markers (c. M) 142. 0

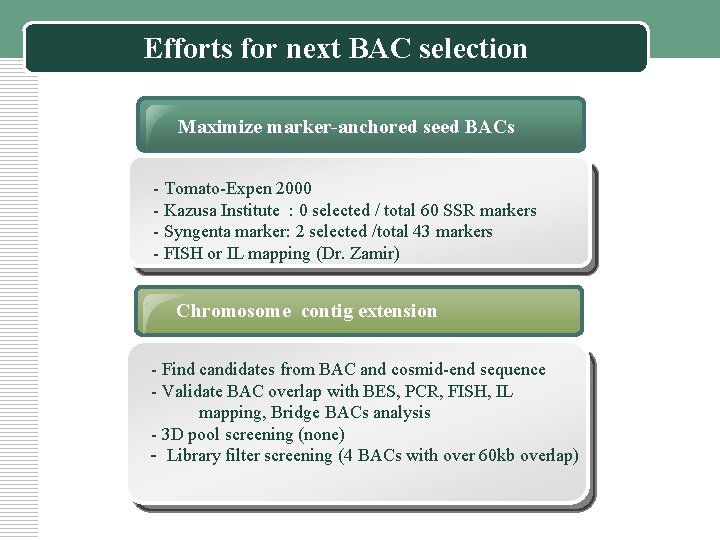
Efforts for next BAC selection Maximize marker-anchored seed BACs - Tomato-Expen 2000 - Kazusa Institute : 0 selected / total 60 SSR markers - Syngenta marker: 2 selected /total 43 markers - FISH or IL mapping (Dr. Zamir) Chromosome contig extension - Find candidates from BAC and cosmid-end sequence - Validate BAC overlap with BES, PCR, FISH, IL mapping, Bridge BACs analysis - 3 D pool screening (none) - Library filter screening (4 BACs with over 60 kb overlap)

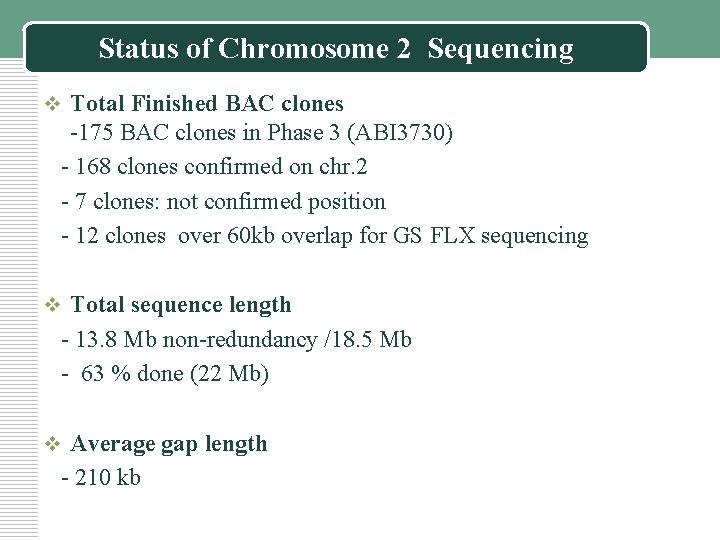
Status of Chromosome 2 Sequencing v Total Finished BAC clones -175 BAC clones in Phase 3 (ABI 3730) - 168 clones confirmed on chr. 2 - 7 clones: not confirmed position - 12 clones over 60 kb overlap for GS FLX sequencing v Total sequence length - 13. 8 Mb non-redundancy /18. 5 Mb - 63 % done (22 Mb) v Average gap length - 210 kb

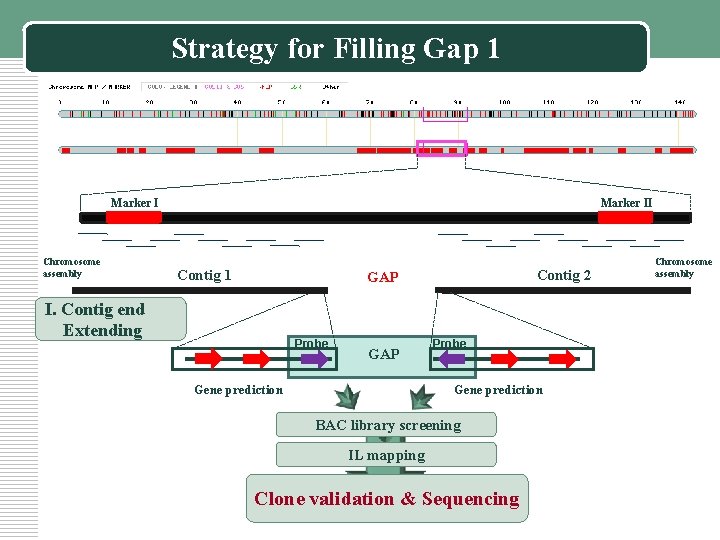
Strategy for Filling Gap 1 Marker I Chromosome assembly Marker II Contig 1 Contig 2 GAP I. Contig end Extending Probe GAP Gene prediction Probe Gene prediction BAC library screening IL mapping Clone validation & Sequencing Chromosome assembly

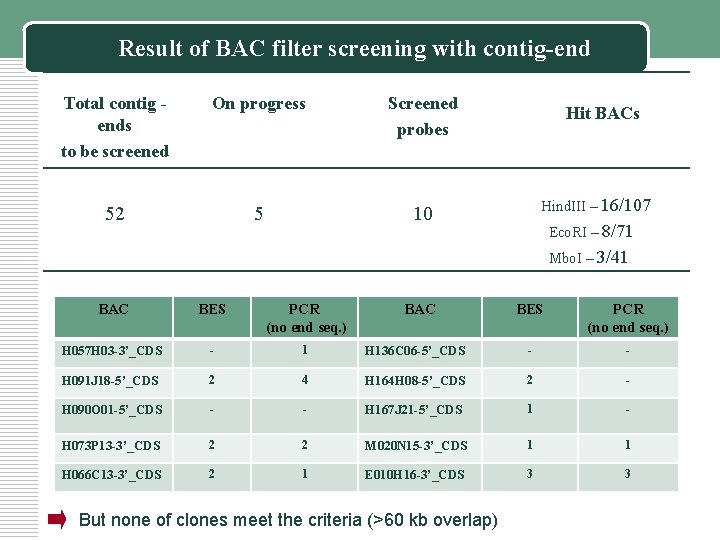
Result of BAC filter screening with contig-end Total contig ends to be screened On progress Screened probes 52 5 10 Hit BACs Hind. III – 16/107 Eco. RI – 8/71 Mbo. I – 3/41 BAC BES PCR (no end seq. ) H 057 H 03 -3’_CDS - 1 H 091 J 18 -5’_CDS 2 H 090 O 01 -5’_CDS BAC BES PCR (no end seq. ) H 136 C 06 -5’_CDS - - 4 H 164 H 08 -5’_CDS 2 - - - H 167 J 21 -5’_CDS 1 - H 073 P 13 -3’_CDS 2 2 M 020 N 15 -3’_CDS 1 1 H 066 C 13 -3’_CDS 2 1 E 010 H 16 -3’_CDS 3 3 But none of clones meet the criteria (>60 kb overlap)

Sequencing of large overlapping clones (>60 kb overlap ) à To make chance to find extendable BAC à GS FLX (~30 X coverage) à BLAST against BES for extension

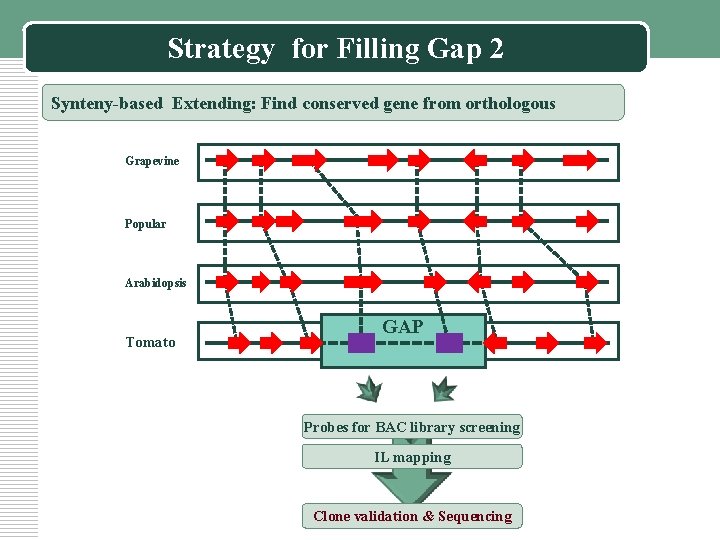
Strategy for Filling Gap 2 Synteny-based Extending: Find conserved gene from orthologous Grapevine Popular Arabidopsis Tomato GAP Probes for BAC library screening IL mapping Clone validation & Sequencing