Using Galaxy for Molecular Assay Design 2012 Galaxy






- Slides: 13

Using Galaxy for Molecular Assay Design 2012 Galaxy Community Conference J Ireland, Principal Bioinformatics Scientist

Galaxy at 5 AM Solutions • 5 AM: Custom software and bioinformatics services company • Galaxy converts for one year o http: //info. 5 amsolutions. com/blog/bid/86705/DIY-Bioinformatics-A -Whole-New-Galaxy • Galaxy in action o Internal bioinformatics resource o Client 1: data source integration o Client 2: custom workflows for assay design o Client 3 (in progress): deploy Galaxy for assay design 2

What is Molecular Assay Design (MAD)? • Design o In-silico process to identify oligos (probes, primers) to detect and/or quantify nucleic acid targets • Balance o Target Coverage: oligo location, expected performance o Cost: $$$, oligo real-estate, oligo interactions • Technologies o PCR, Taq. Man, molecular inversion probes, LNA, oligo arrays, … • Applications o Targeted sequencing, genotyping, gene expression, mi. RNA, … 3

The MAD Workflow • Target Selection o Identify locations on genome/transcriptome of interest while avoiding trouble regions • Assay Design o Select sets of primers and/or probes against targets • Assay Prioritization/Filter o Filter, prioritize and annotate assay designs • Ensemble Selection o Select final combination(s) of assays 4

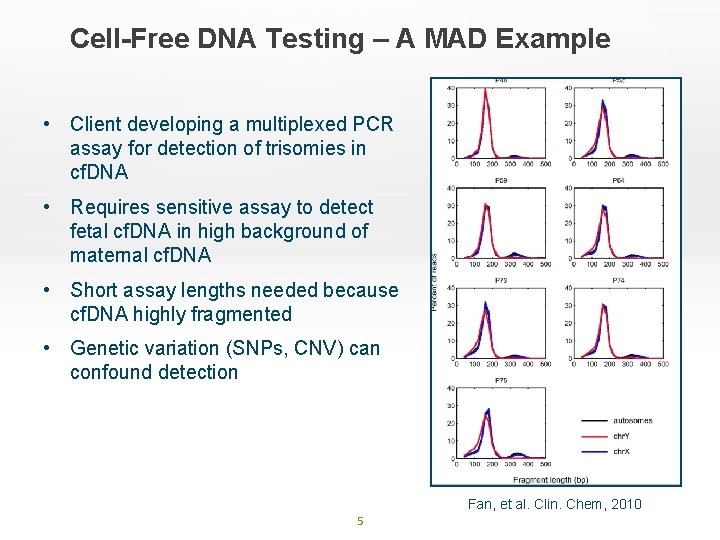
Cell-Free DNA Testing – A MAD Example • Client developing a multiplexed PCR assay for detection of trisomies in cf. DNA • Requires sensitive assay to detect fetal cf. DNA in high background of maternal cf. DNA • Short assay lengths needed because cf. DNA highly fragmented • Genetic variation (SNPs, CNV) can confound detection Fan, et al. Clin. Chem, 2010 5

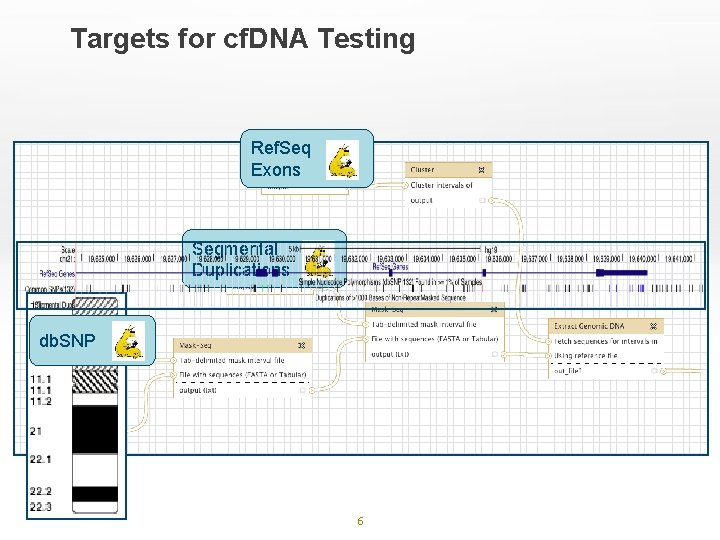
Targets for cf. DNA Testing Ref. Seq Exons Segmental Duplications db. SNP 6

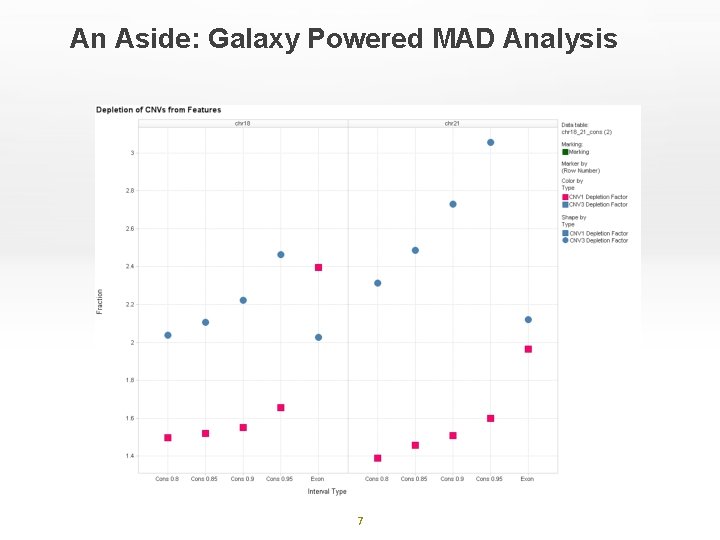
An Aside: Galaxy Powered MAD Analysis 7

Assay Design 8

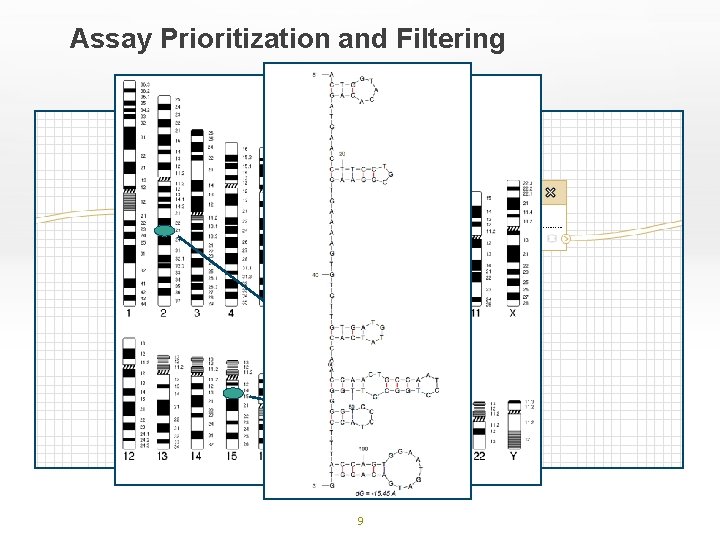
Assay Prioritization and Filtering 9

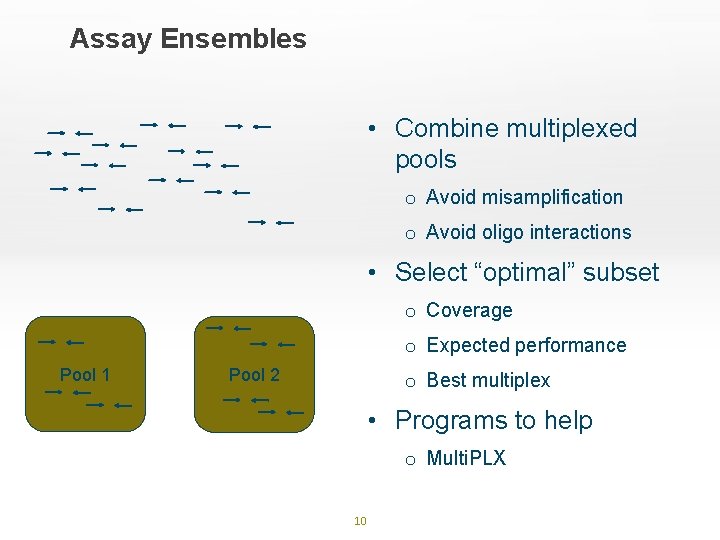
Assay Ensembles • Combine multiplexed pools o Avoid misamplification o Avoid oligo interactions • Select “optimal” subset o Coverage o Expected performance Pool 1 Pool 2 o Best multiplex • Programs to help o Multi. PLX 10

Looking Back • No more Perl soup • Data sets, parameters, steps preserved • Reproducible design • Sharable with client 11

Looking Ahead • Tool Shed submission • Extend workflows to ensemble selection • Automate report generation • Sweave for Galaxy? 12

Thanks! Andy Evans Will Fitz. Hugh The Galaxy Community! 13