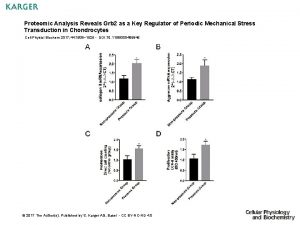
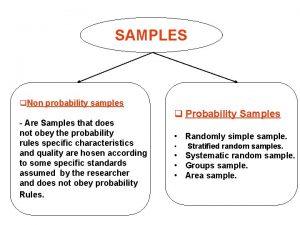
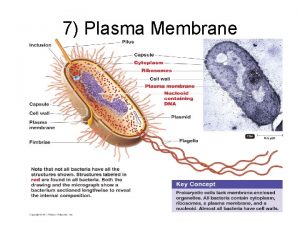
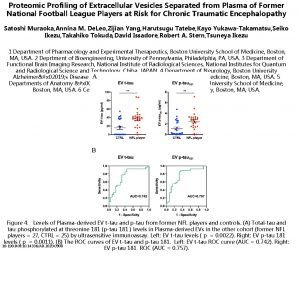
Tissueenhanced proteomic analysis of plasma samples reveals new





- Slides: 20

Tissue-enhanced proteomic analysis of plasma samples reveals new mechanistic biomarker candidates for the stratification of Amyotrophic lateral sclerosis patients. Irene Zubiri MNDA Symposium 2016 i. zubiri@qmul. ac. uk

Summary • Phenotypic variability in ALS. • Animal models mimicking human ALS. • The urgent need of new biomarkers. • TMT LC-MS/MS and TMTcalibrator™. • Ongoing studies and Results: Tissue biomarkers in human plasma samples and in an equivalent study in a mouse model. • Future perspectives and Conclusions.

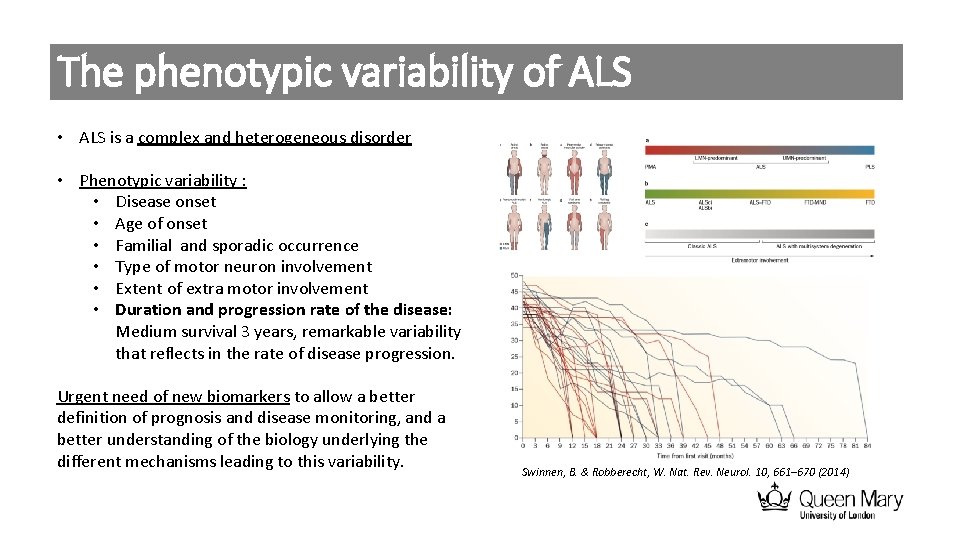
The phenotypic variability of ALS • ALS is a complex and heterogeneous disorder • Phenotypic variability : • Disease onset • Age of onset • Familial and sporadic occurrence • Type of motor neuron involvement • Extent of extra motor involvement • Duration and progression rate of the disease: Medium survival 3 years, remarkable variability that reflects in the rate of disease progression. Urgent need of new biomarkers to allow a better definition of prognosis and disease monitoring, and a better understanding of the biology underlying the different mechanisms leading to this variability. Swinnen, B. & Robberecht, W. Nat. Rev. Neurol. 10, 661– 670 (2014)

The use of animal models • The development of rodent models of ALS recapitulating the main aspects of human pathology has been a game changing event. • The reliance on animal models of the disease in support of clinical trials has not been particularly rewarding. • Defining similarities and differences between the animal models and the human pathologies is key to the discovery of new biomarkers. Many drugs giving positive results in animal models have failed to shift disease progression in humans. Does the pathobiology of ALS in animals fully reflect that of affected humans?

Biomarker Sources “ An objective measurement that acts as an indicator of normal biological processes, pathogenic processes or pharmacologic responses to therapeutic intervention”. Untargeted proteomic mass spectrometry techniques. Protein levels

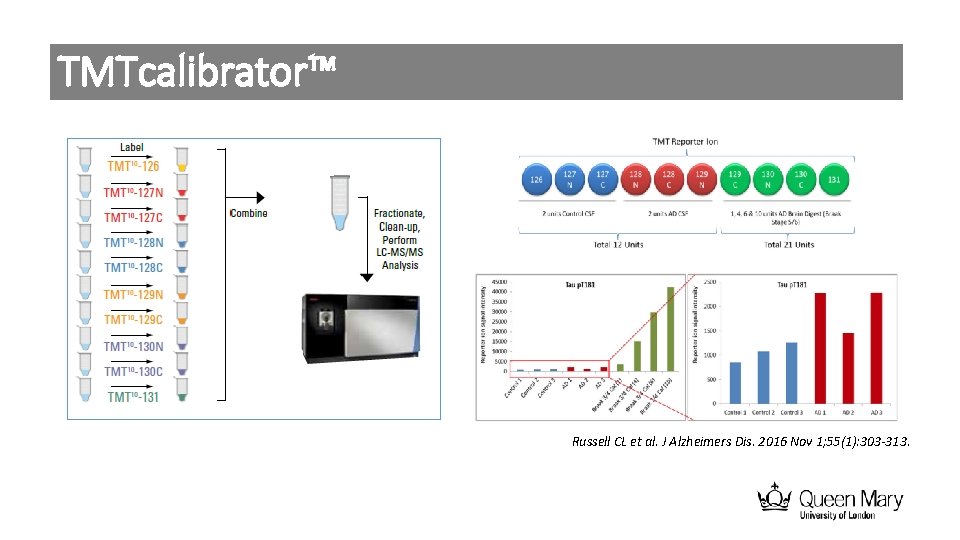
TMTcalibrator™ Russell CL et al. J Alzheimers Dis. 2016 Nov 1; 55(1): 303 -313.

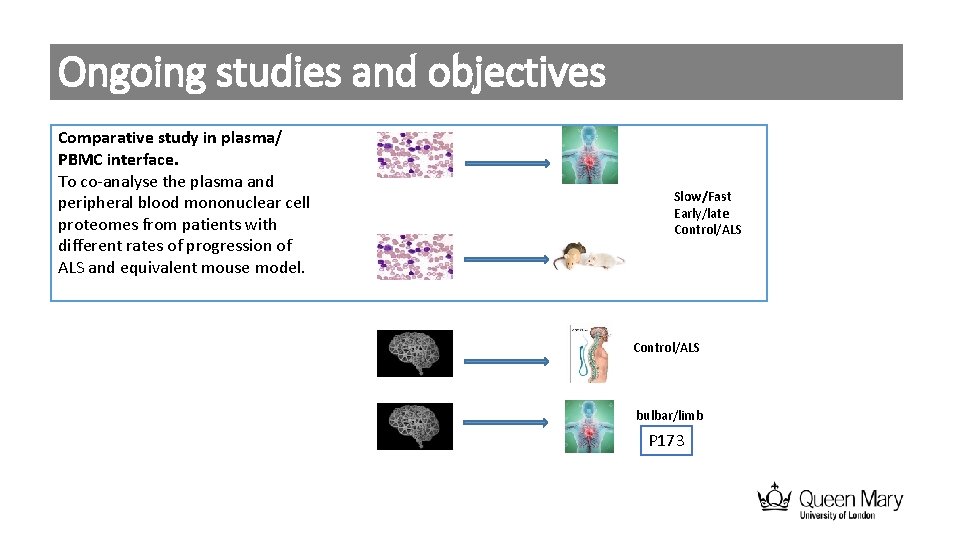
Ongoing studies and objectives Comparative study in plasma/ PBMC interface. To co-analyse the plasma and peripheral blood mononuclear cell proteomes from patients with different rates of progression of ALS and equivalent mouse model. Slow/Fast Early/late Control/ALS bulbar/limb P 173

Plasma/PBMC study in human Samples Reference: Pool of PBMCs Representing all the conditions under study

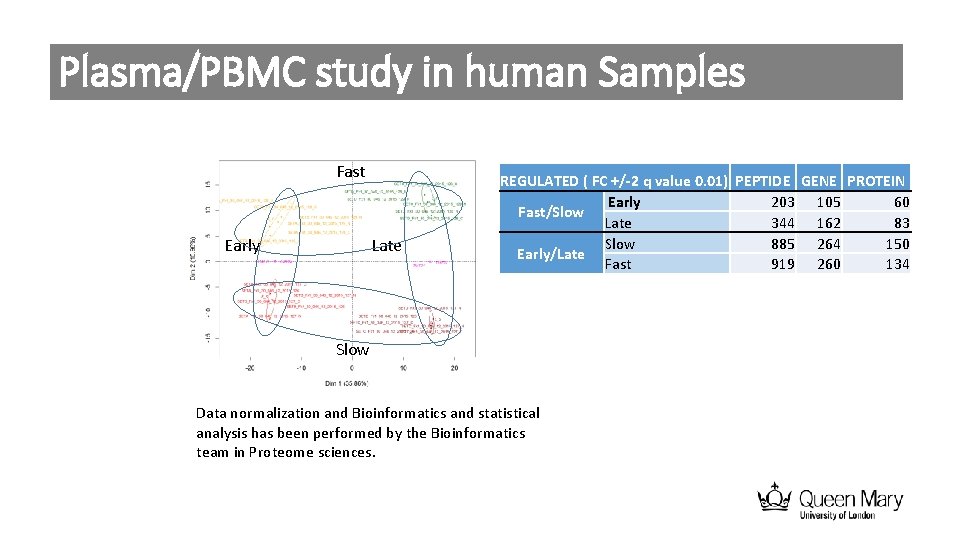
Plasma/PBMC study in human Samples Fast Early Late REGULATED ( FC +/-2 q value 0. 01) PEPTIDE GENE PROTEIN Early 203 105 60 Fast/Slow Late 344 162 83 Slow 885 264 150 Early/Late Fast 919 260 134 Slow Data normalization and Bioinformatics and statistical analysis has been performed by the Bioinformatics team in Proteome sciences.

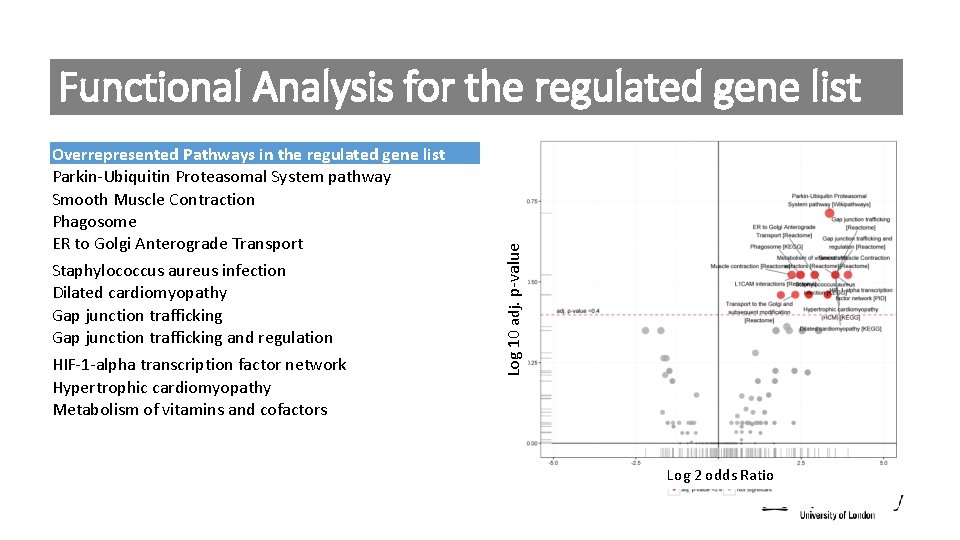
Overrepresented Pathways in the regulated gene list Parkin-Ubiquitin Proteasomal System pathway Smooth Muscle Contraction Phagosome ER to Golgi Anterograde Transport Staphylococcus aureus infection Dilated cardiomyopathy Gap junction trafficking and regulation HIF-1 -alpha transcription factor network Hypertrophic cardiomyopathy Metabolism of vitamins and cofactors Log 10 adj. p-value Functional Analysis for the regulated gene list Log 2 odds Ratio

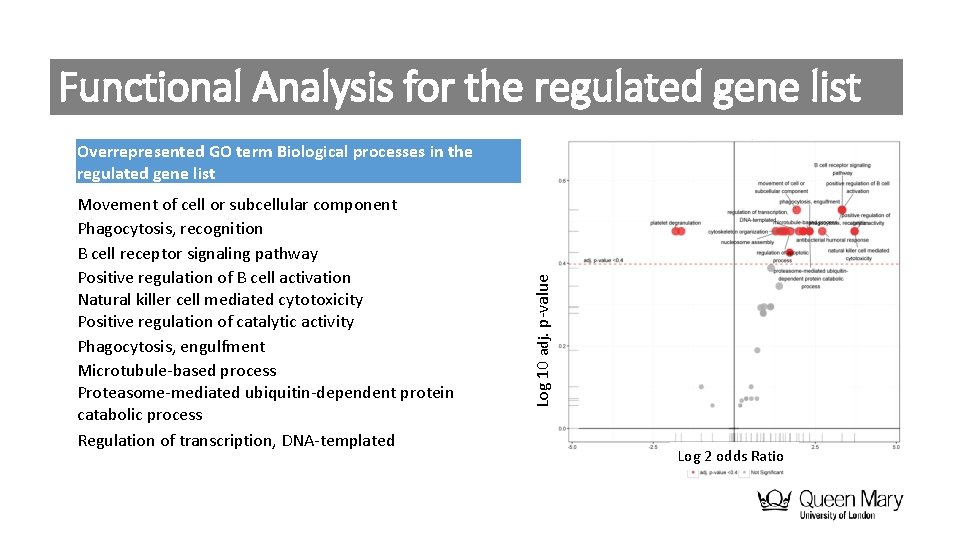
Functional Analysis for the regulated gene list Movement of cell or subcellular component Phagocytosis, recognition B cell receptor signaling pathway Positive regulation of B cell activation Natural killer cell mediated cytotoxicity Positive regulation of catalytic activity Phagocytosis, engulfment Microtubule-based process Proteasome-mediated ubiquitin-dependent protein catabolic process Regulation of transcription, DNA-templated Log 10 adj. p-value Overrepresented GO term Biological processes in the regulated gene list Log 2 odds Ratio

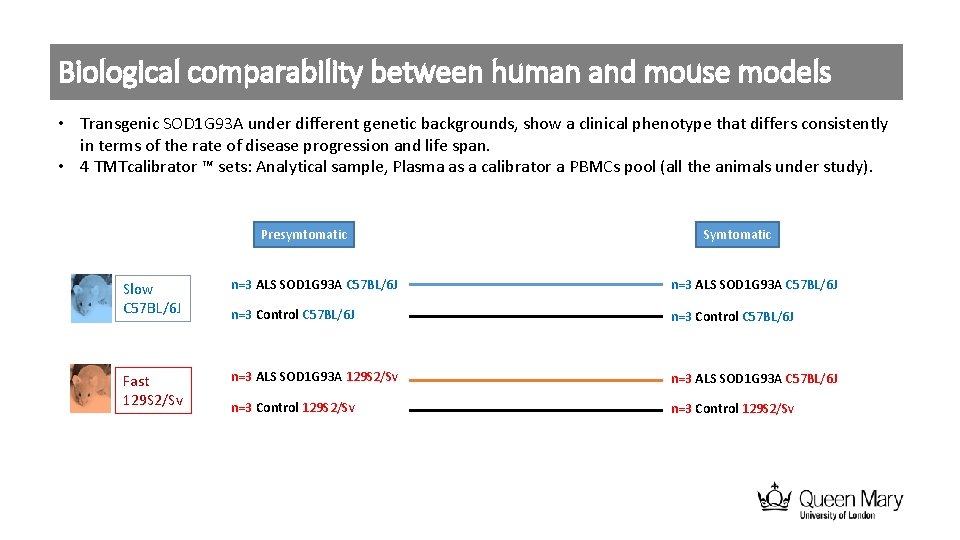
Biological comparability between human and mouse models • Transgenic SOD 1 G 93 A under different genetic backgrounds, show a clinical phenotype that differs consistently in terms of the rate of disease progression and life span. • 4 TMTcalibrator ™ sets: Analytical sample, Plasma as a calibrator a PBMCs pool (all the animals under study). Presymtomatic Slow C 57 BL/6 J n=3 ALS SOD 1 G 93 A C 57 BL/6 J n=3 Control C 57 BL/6 J Fast 129 S 2/Sv n=3 ALS SOD 1 G 93 A C 57 BL/6 J n=3 Control 129 S 2/Sv

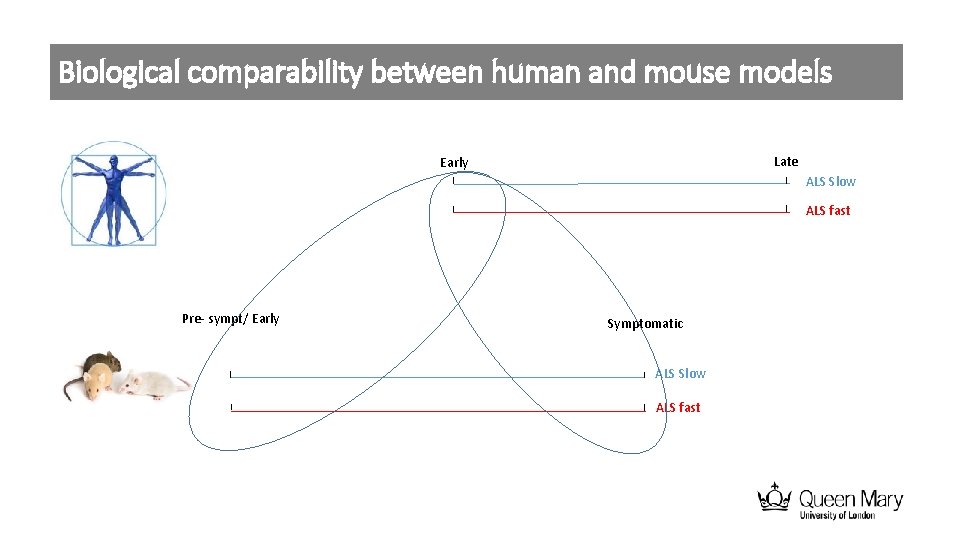
Biological comparability between human and mouse models Late Early ALS Slow ALS fast Pre- sympt/ Early Symptomatic ALS Slow ALS fast

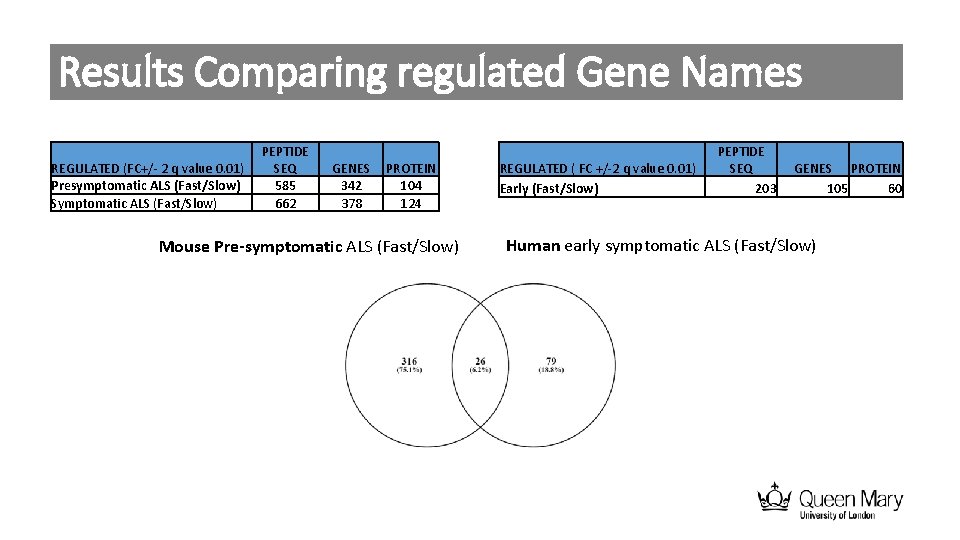
Results Comparing regulated Gene Names REGULATED (FC+/- 2 q value 0. 01) Presymptomatic ALS (Fast/Slow) Symptomatic ALS (Fast/Slow) PEPTIDE SEQ 585 662 GENES 342 378 PROTEIN 104 124 Mouse Pre-symptomatic ALS (Fast/Slow) REGULATED ( FC +/-2 q value 0. 01) Early (Fast/Slow) PEPTIDE SEQ 203 GENES PROTEIN 105 60 Human early symptomatic ALS (Fast/Slow)

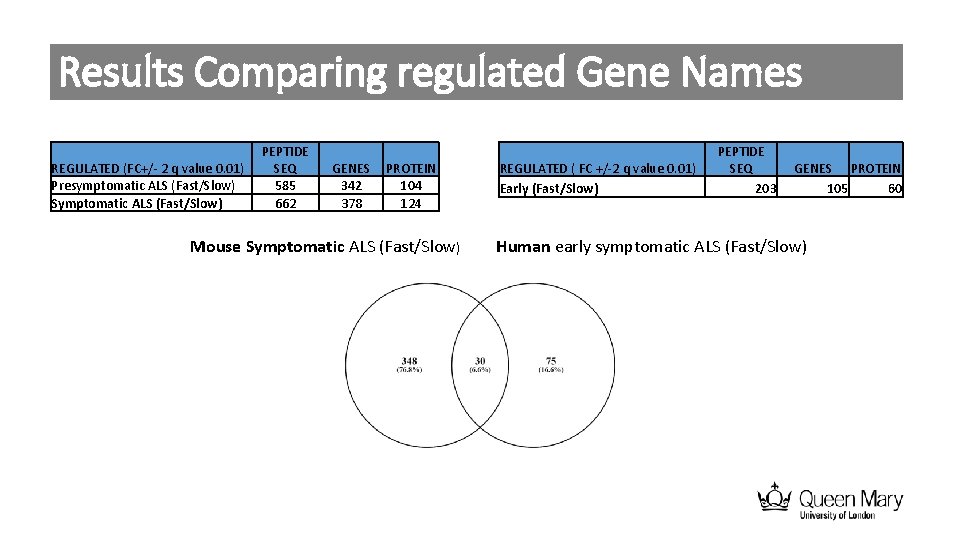
Results Comparing regulated Gene Names REGULATED (FC+/- 2 q value 0. 01) Presymptomatic ALS (Fast/Slow) Symptomatic ALS (Fast/Slow) PEPTIDE SEQ 585 662 GENES 342 378 PROTEIN 104 124 Mouse Symptomatic ALS (Fast/Slow) REGULATED ( FC +/-2 q value 0. 01) Early (Fast/Slow) PEPTIDE SEQ 203 GENES PROTEIN 105 60 Human early symptomatic ALS (Fast/Slow)

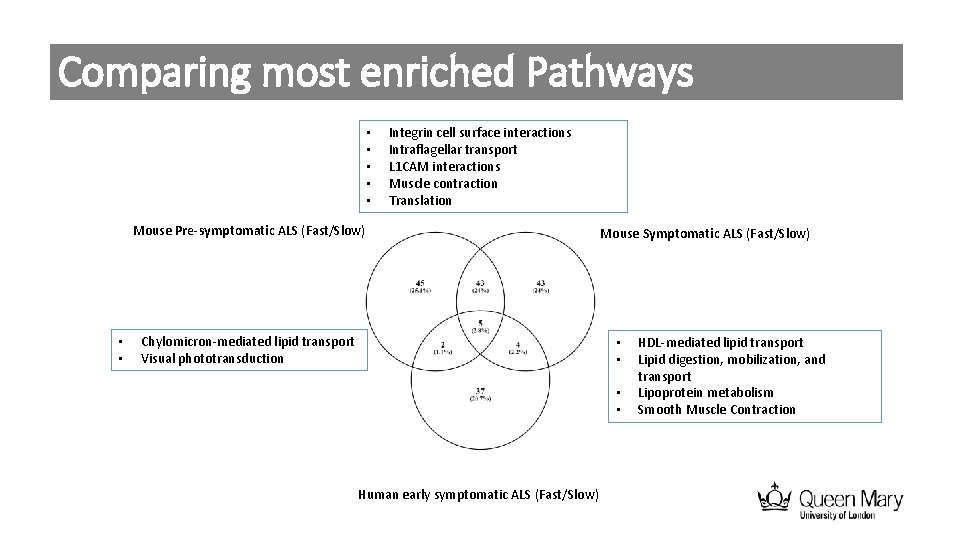
Comparing most enriched Pathways • • • Integrin cell surface interactions Intraflagellar transport L 1 CAM interactions Muscle contraction Translation Mouse Pre-symptomatic ALS (Fast/Slow) • • Chylomicron-mediated lipid transport Visual phototransduction Mouse Symptomatic ALS (Fast/Slow) • • Human early symptomatic ALS (Fast/Slow) HDL-mediated lipid transport Lipid digestion, mobilization, and transport Lipoprotein metabolism Smooth Muscle Contraction

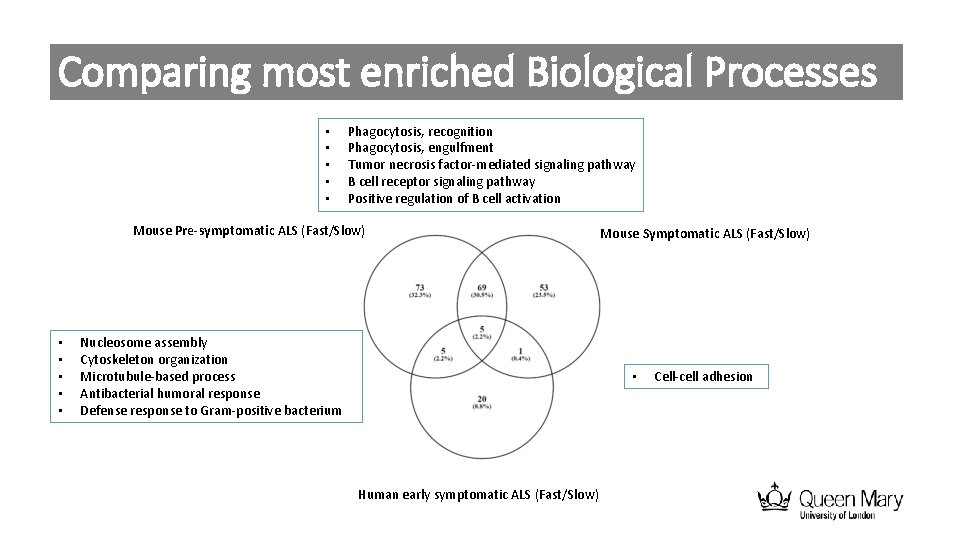
Comparing most enriched Biological Processes • • • Phagocytosis, recognition Phagocytosis, engulfment Tumor necrosis factor-mediated signaling pathway B cell receptor signaling pathway Positive regulation of B cell activation Mouse Pre-symptomatic ALS (Fast/Slow) • • • Nucleosome assembly Cytoskeleton organization Microtubule-based process Antibacterial humoral response Defense response to Gram-positive bacterium Mouse Symptomatic ALS (Fast/Slow) • Human early symptomatic ALS (Fast/Slow) Cell-cell adhesion

Future perspective • Validation of the most reliable biomarker candidates using orthogonal techniques. Best potential biomarkers: regulated in both studies and relevant within the overlapping pathways and biological functions. • A parallel data analysis is being carried out for the mouse model comparing Control/ ALS at each time point and longitudinally. The results of that analysis are being confronted with the longitudinal data derived from the human study. To characterize good biomarkers for diagnosis and monitoring.

Conclusions • The TMTcalibrator™ technique has provided a unique opportunity to study the interface between cellular and fluid components of blood in unravelling of ALS pathology. • The parallel analysis of SOD 1 mouse models provided a formidable working platform with translational potential. • A large database has been generated and will be soon publicly available to support other researchers working on ALS. • The validation of some of the most relevant biomarker candidates will represent a promising biomarker panel: • A better Patient Stratification: to better predict the disease progression and to make a more informed decision about the inclusion of patients in clinical trials. • Selection of better drug target to be tested in mouse models.

Acknowledgements • The Patients and their families. • Academic and Industrial support: • QMUL, Blizard Institute • Andrea Malaspina • Ching-Hua Lu • Rocco Adiutori • Vittoria Lombardi • Johan Aarum • Denise Sheer • UCL Institute of Neurology. • Linda Greensmith • Mario Negri Institute for Pharmacological Research: • Caterina Bendotti • Giovanni Nardo • Proteome sciences London • Ian Pike • Malcolm Ward • Michael Bremang • Vikram Mitra • Emanuela Leoni • Richard Churchus • Eva Sedlak • Claire Russell • Hui-Chung Liang • Proteome Sciences Frankfurt • Karsten Khun • Gitte Boehm • Sasa Koncarevic i. zubiri@qmul. ac. uk • Economic support:
 Is theme and main idea the same thing
Is theme and main idea the same thing Art reveals society
Art reveals society Mark twain reveals stage fright
Mark twain reveals stage fright Indirect and direct characterization
Indirect and direct characterization Theme of dulce et decorum est
Theme of dulce et decorum est The theme is the central idea or truth in the story
The theme is the central idea or truth in the story A sustainability report reveals a firm's
A sustainability report reveals a firm's Narrative writing reveals something of importance.
Narrative writing reveals something of importance. Which narration reveals a character with a shy personality?
Which narration reveals a character with a shy personality? Who reveals that his supporters will be made earls
Who reveals that his supporters will be made earls Act 4 scene 6 hamlet summary
Act 4 scene 6 hamlet summary A good author reveals a character through
A good author reveals a character through Tea telpas writing prompts
Tea telpas writing prompts Event samples observations
Event samples observations Formula for t test independent samples
Formula for t test independent samples Dependent samples
Dependent samples Dependent t-test
Dependent t-test A dietitian wishes to see if a person's cholesterol
A dietitian wishes to see if a person's cholesterol Successive independent samples design
Successive independent samples design Write an opinion paragraph
Write an opinion paragraph Dependent ttest
Dependent ttest