Simple Lectin Identification Using a Chemical Tongue Lucienne

- Slides: 23

Simple Lectin Identification Using a ‘Chemical Tongue’ Lucienne Otten Supervised by: Dr. Matthew Gibson

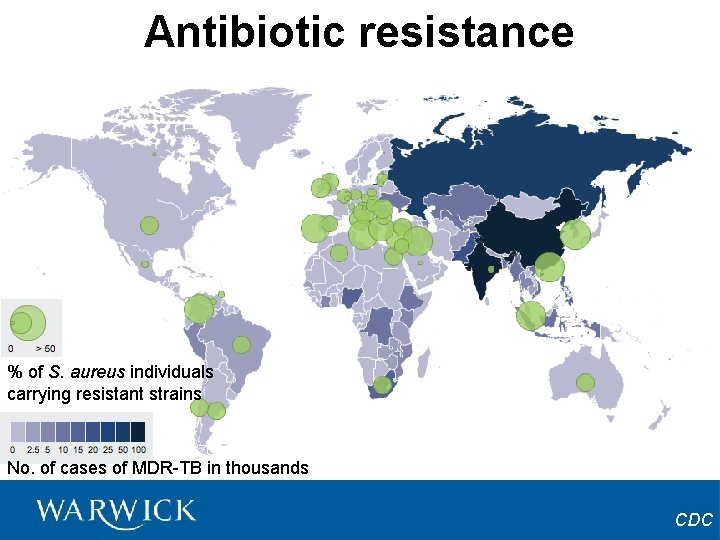
Antibiotic resistance % of S. aureus individuals carrying resistant strains No. of cases of MDR-TB in thousands CDC

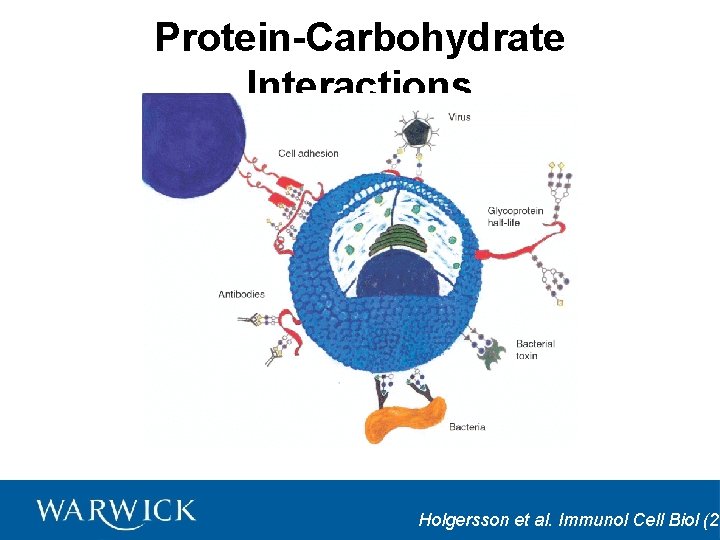
Protein-Carbohydrate Interactions Holgersson et al. Immunol Cell Biol (20

Point of care diagnostics CFG database

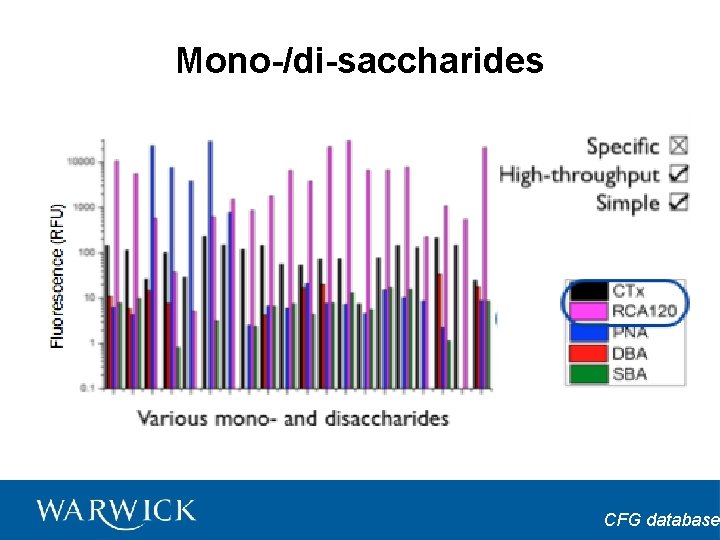
Mono-/di-saccharides CFG database

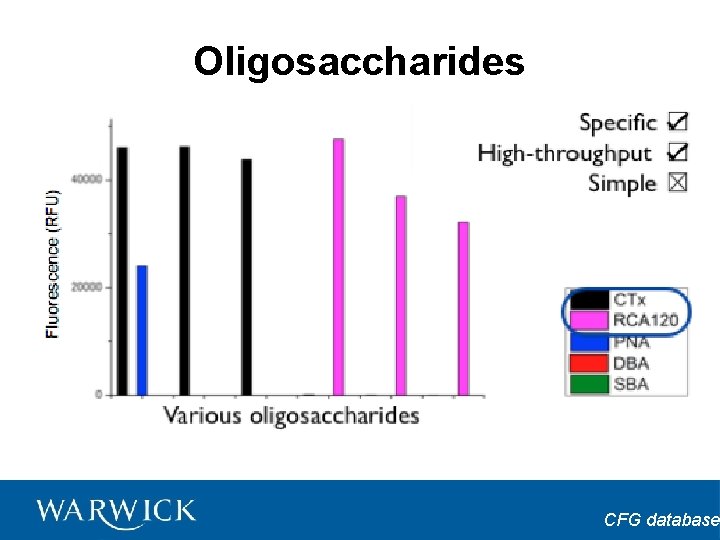
Oligosaccharides CFG database

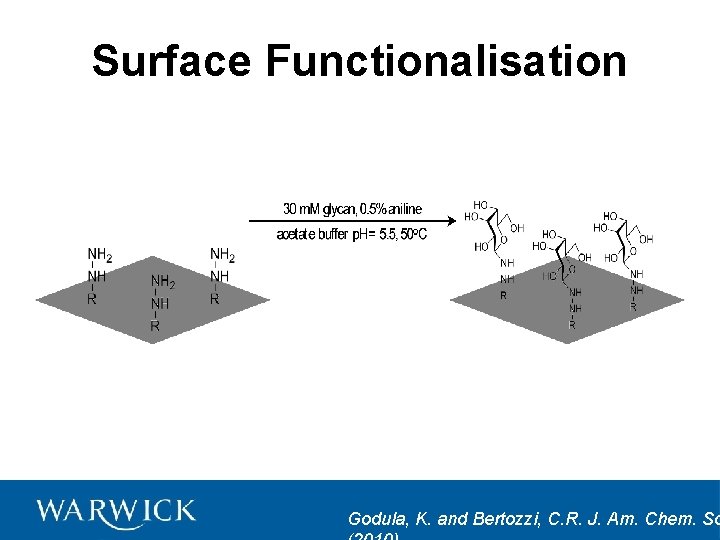
Surface Functionalisation Godula, K. and Bertozzi, C. R. J. Am. Chem. So

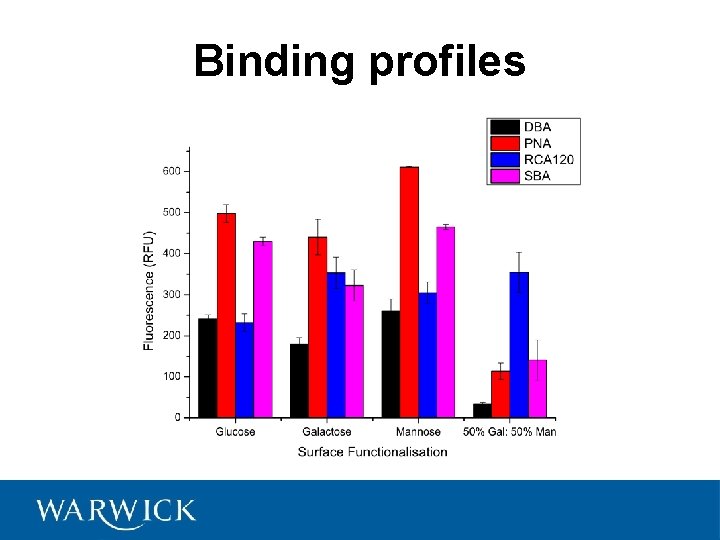
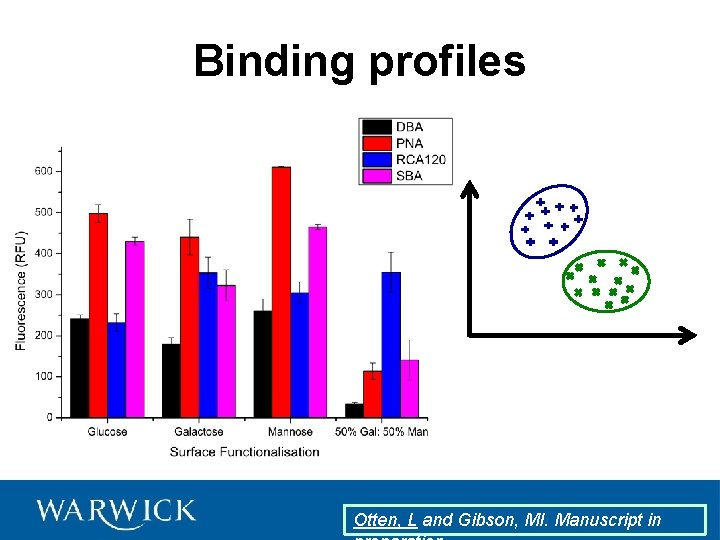
Binding profiles

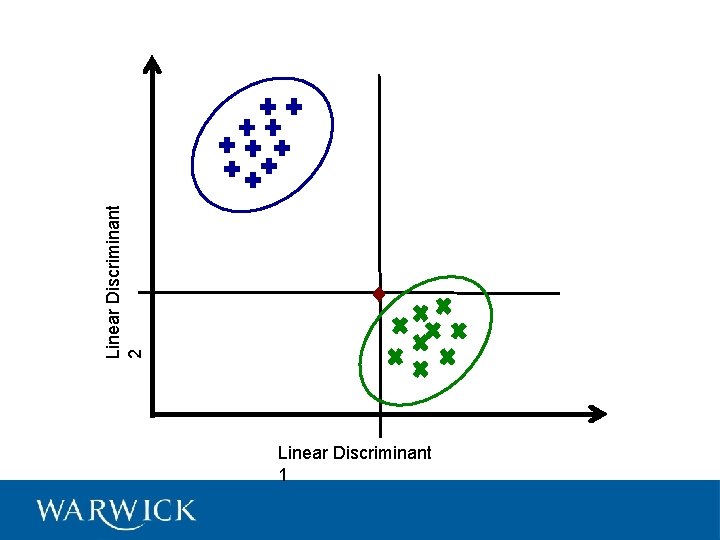
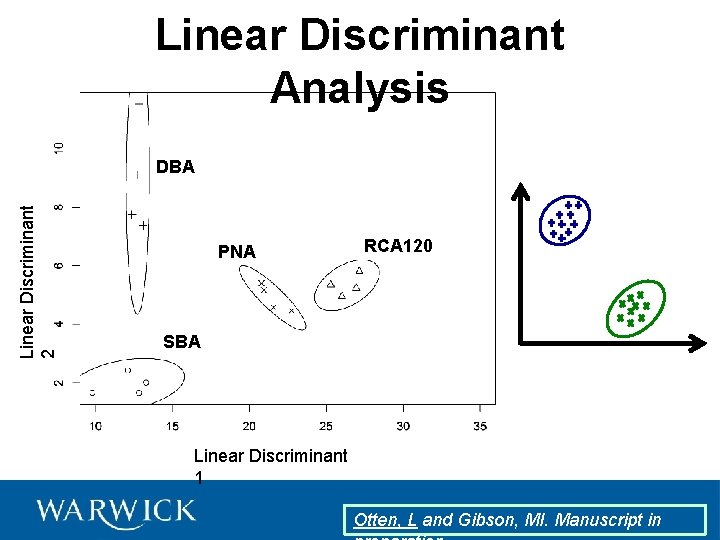
Linear Discriminant Analysis

Linear Discriminant 2 Linear Discriminant 1

Binding profiles Otten, L and Gibson, MI. Manuscript in

Linear Discriminant Analysis Linear Discriminant 2 DBA PNA RCA 120 SBA Linear Discriminant 1 Otten, L and Gibson, MI. Manuscript in

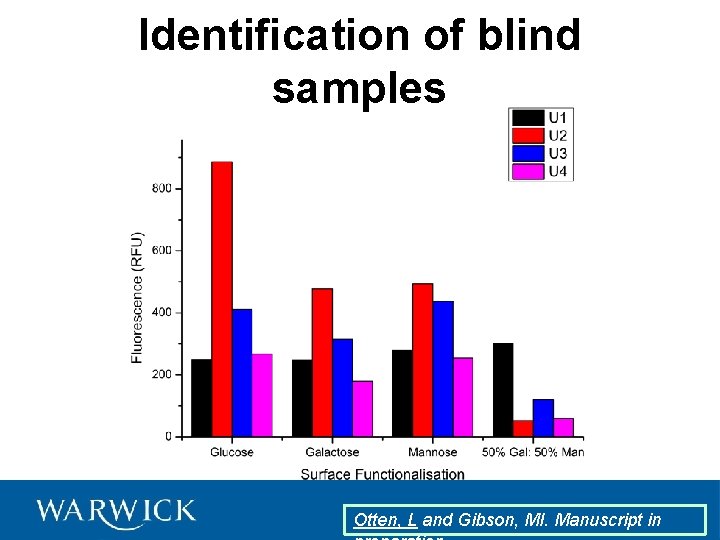
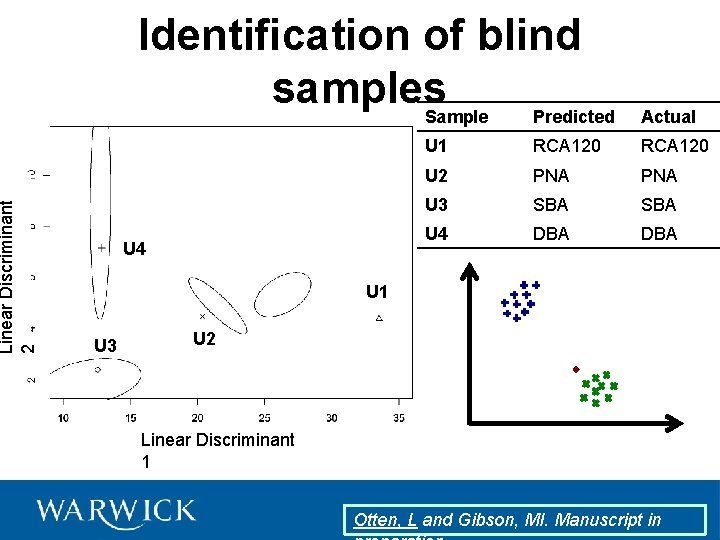
Identification of blind samples Otten, L and Gibson, MI. Manuscript in

Linear Discriminant 2 Identification of blind samples Sample Predicted U 4 Actual U 1 RCA 120 U 2 PNA U 3 SBA U 4 DBA U 1 U 3 U 2 Linear Discriminant 1 Otten, L and Gibson, MI. Manuscript in

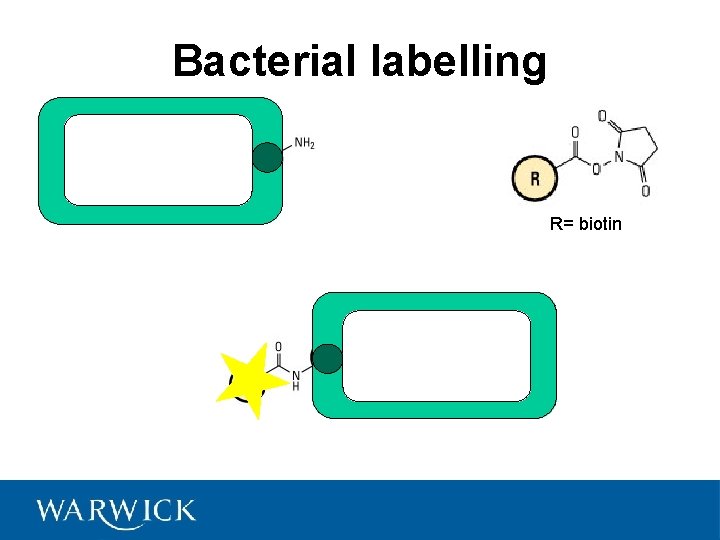
Bacterial labelling R= biotin

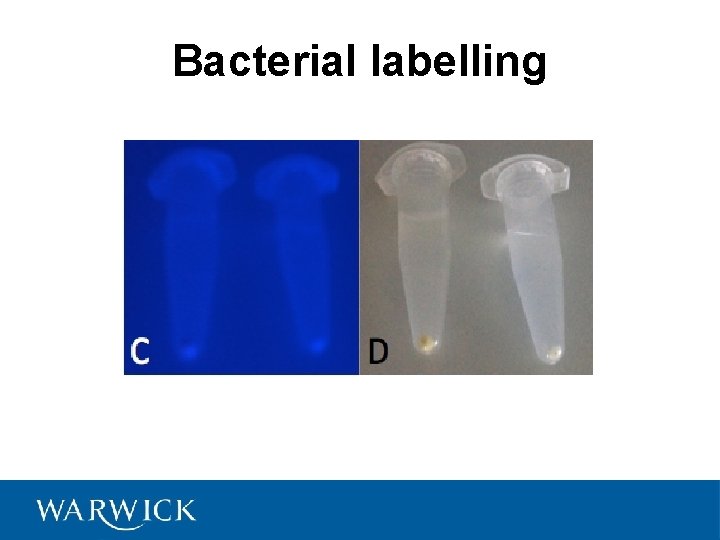
Bacterial labelling

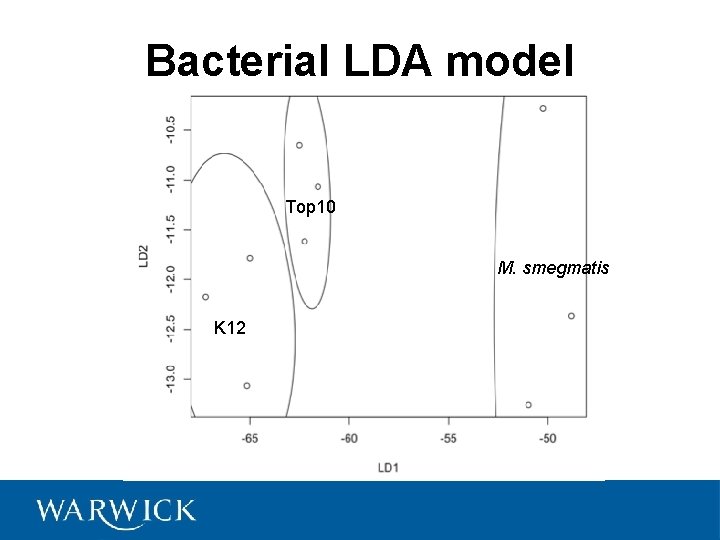
Bacterial LDA model Top 10 M. smegmatis K 12

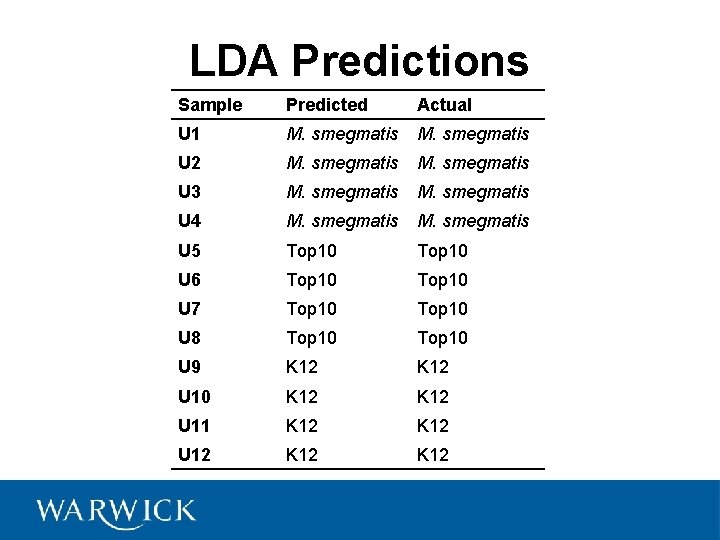
LDA Predictions Sample Predicted Actual U 1 M. smegmatis U 2 M. smegmatis U 3 M. smegmatis U 4 M. smegmatis U 5 Top 10 U 6 Top 10 U 7 Top 10 U 8 Top 10 U 9 K 12 U 10 K 12 U 11 K 12 U 12 K 12

CFG data Fim. H Fli. D Typhoid toxin CFG database

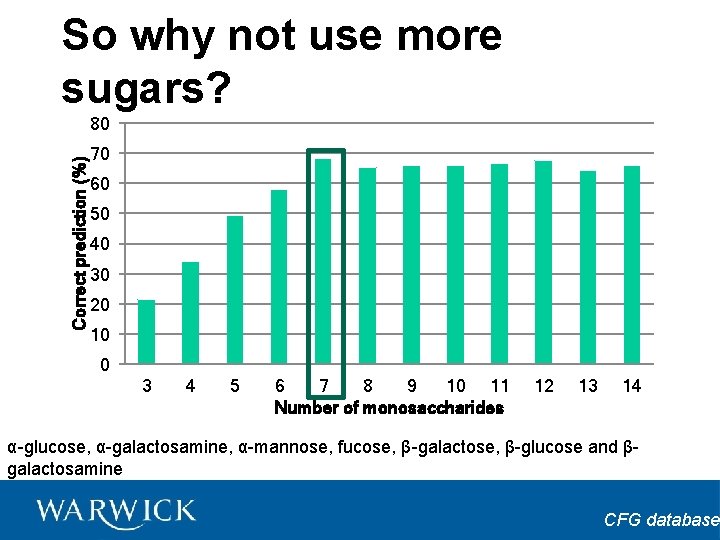
So why not use more sugars? Correct prediction (%) 80 70 60 50 40 30 20 10 0 3 4 5 6 7 8 9 10 11 Number of monosaccharides 12 13 14 α-glucose, α-galactosamine, α-mannose, fucose, β-galactose, β-glucose and βgalactosamine CFG database

Conclusions • Linear discriminant analysis has been shown to aid in the identification of blind lectin samples • It has also shown to be applicable to identification of bacterial species • Analysis of larger data sets highlighted the need for identifying ‘optimum surfaces’.

Future work • Extend the bacterial LDA analysis to more species of bacteria • Extend the LDA analysis of the CFG database to beyond monosaccharides.

Acknowledgements Gibson Group Members - Gemma-Louise Davies Yunhua Chen Catherine Cooper Robert Deller - - Caroline Biggs Daniel Phillips Tom Congdon Sarah-Jane Richards - Sang-Ho Won - Laura Wilkins - Robert Keogh Daniel Mitchell Richard Lowery Ben Martyn Lewis Blackman Matthew Gibson