Nutrigenomicspharmacogenomics Lactose intolerance CT13910 lactase persistencenon functions in
- Slides: 9

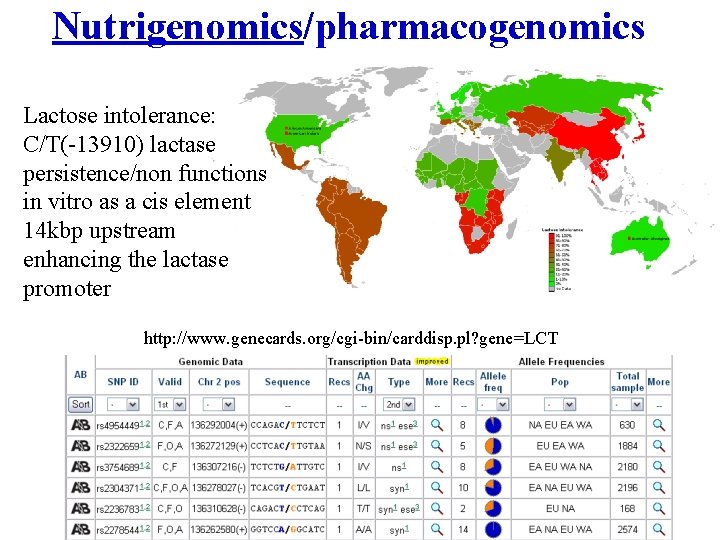
Nutrigenomics/pharmacogenomics Lactose intolerance: C/T(-13910) lactase persistence/non functions in vitro as a cis element 14 kbp upstream enhancing the lactase promoter http: //www. genecards. org/cgi-bin/carddisp. pl? gene=LCT

Nutrigenomics/pharmacogenomics Thiopurine methyltransferase (TPMT) metabolizes 6 mercaptopurine and azathiopurine, two drugs used in a range of indications, from childhood leukemia to autoimmune diseases CYP 450 superfamily: CYP 2 D 6 has over 75 known allelic variations, 30% of people in parts of East Africa have multiple copies of the gene, not be adequately treated with standard doses of drugs, e. g. codeine (activated by CYP 2 D 6).

Further optimization readings Duarte et al. reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007 Feb 6; 104(6): 1777 -82. Joyce AR, Palsson BO. Toward whole cell modeling and simulation: comprehensive functional genomics through the constraint-based approach. Prog Drug Res. 2007; 64: 265, 267 -309. Review. Herring, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006 38: 1406 -12. Desai RP, Nielsen LK, Papoutsakis ET. Stoichiometric modeling of Clostridium acetobutylicum fermentations with non-linear constraints. J Biotechnol. 1999 71: 191 -205.

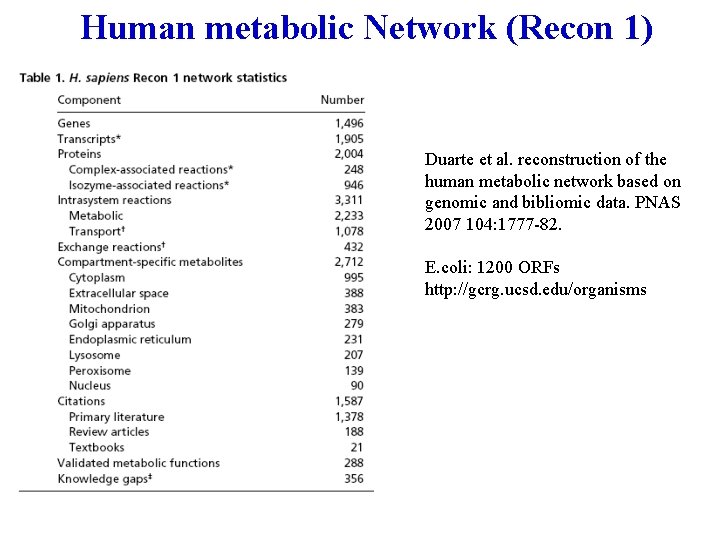
Human metabolic Network (Recon 1) Duarte et al. reconstruction of the human metabolic network based on genomic and bibliomic data. PNAS 2007 104: 1777 -82. E. coli: 1200 ORFs http: //gcrg. ucsd. edu/organisms

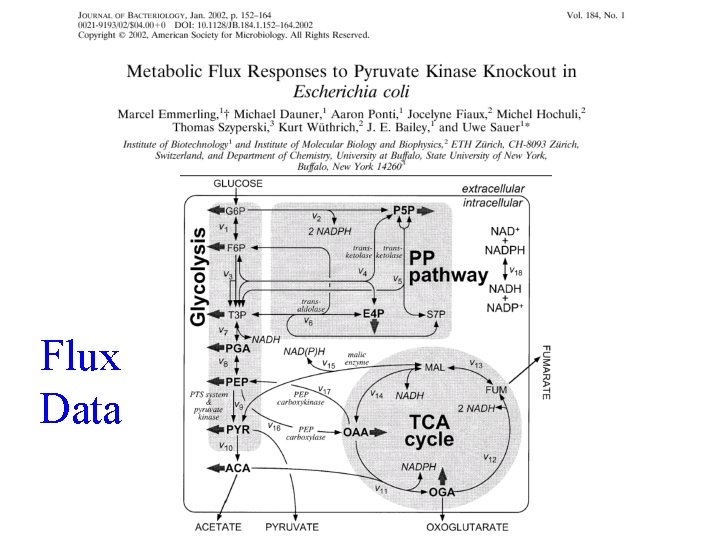
Flux Data

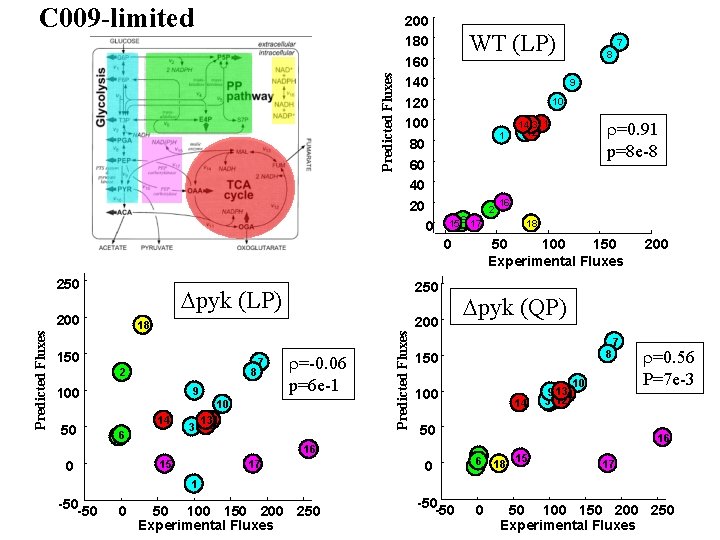
Predicted Fluxes C 009 -limited 200 180 160 140 120 100 80 60 40 20 0 WT (LP) 9 10 1 6 17 1545 0 250 18 150 8 2 7 9 100 14 5 46 3 r=-0. 06 p=6 e-1 10 13 11 12 Predicted Fluxes 200 50 250 Dpyk (LP) 200 15 17 2 141311 312 r=0. 91 p=8 e-8 16 18 50 100 150 Experimental Fluxes 8 150 100 14 10 9 13 11 31 12 50 0 200 Dpyk (QP) 7 16 0 7 8 r=0. 56 P=7 e-3 16 15 62 5 4 18 17 1 -50 -50 0 50 100 150 200 250 Experimental Fluxes

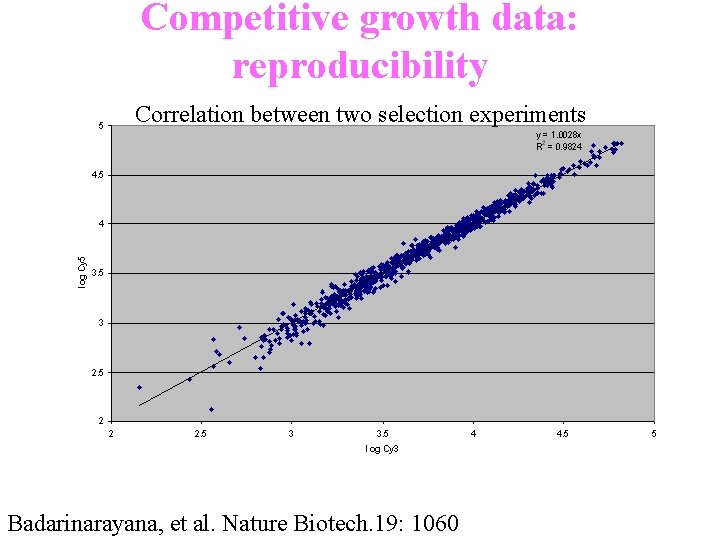
Competitive growth data: reproducibility Correlation between two selection experiments Badarinarayana, et al. Nature Biotech. 19: 1060

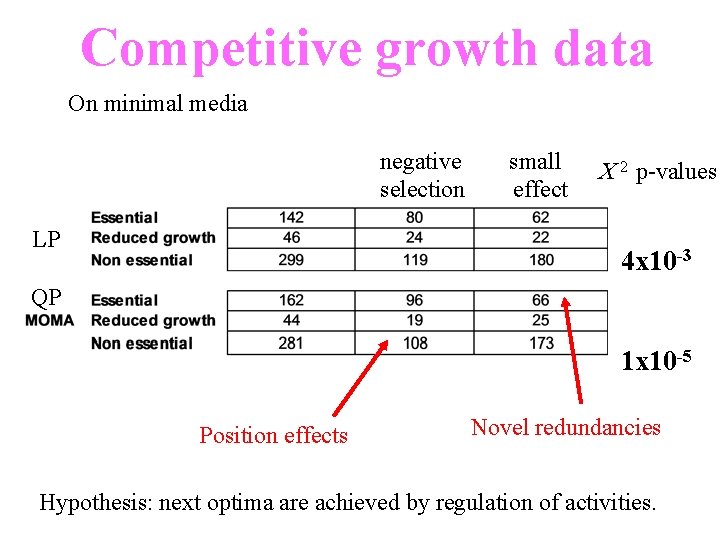
Competitive growth data On minimal media negative selection LP small effect C 2 p-values 4 x 10 -3 QP 1 x 10 -5 Position effects Novel redundancies Hypothesis: next optima are achieved by regulation of activities.

Intelligent Design & Metabolic Evolution Fong SS, Burgard AP, Herring CD, Knight EM, Blattner FR, Maranas CD, Palsson BO. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng. 2005 91(5): 643 -8. Rozen DE, Schneider D, Lenski RE Long-term experimental evolution in Escherichia coli. XIII. Phylogenetic history of a balanced polymorphism. J Mol Evol. 2005 61(2): 171 -80 Andries K, et al. (J&J) A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005 307: 223 -7. Shendure et al. Accurate Multiplex Polony Sequencing of an Evolved Bacterial Genome Science 2005 309: 1728 (Select for secretion & ‘altruism’).