Identifying and classifying functional small RNAs from pine




- Slides: 19

Identifying and classifying functional small RNAs from pine Ryan Morin BC Genome Sciences Centre (presenting research conducted in the lab of Dr. Peter Unrau)

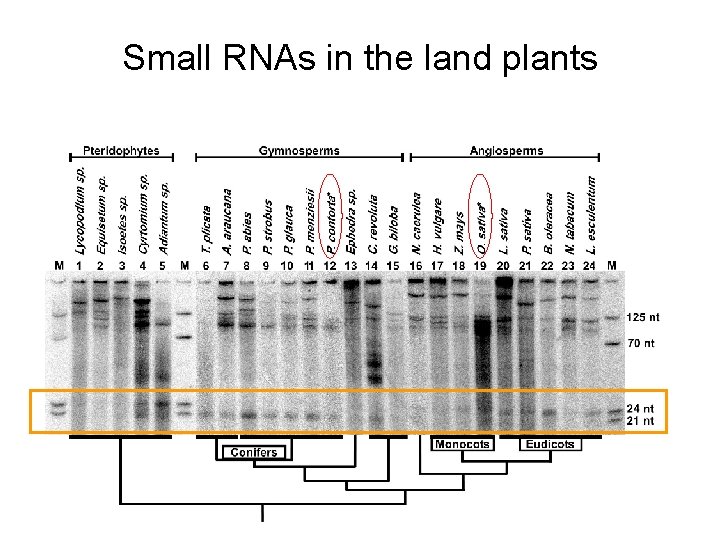
Small RNAs in the land plants

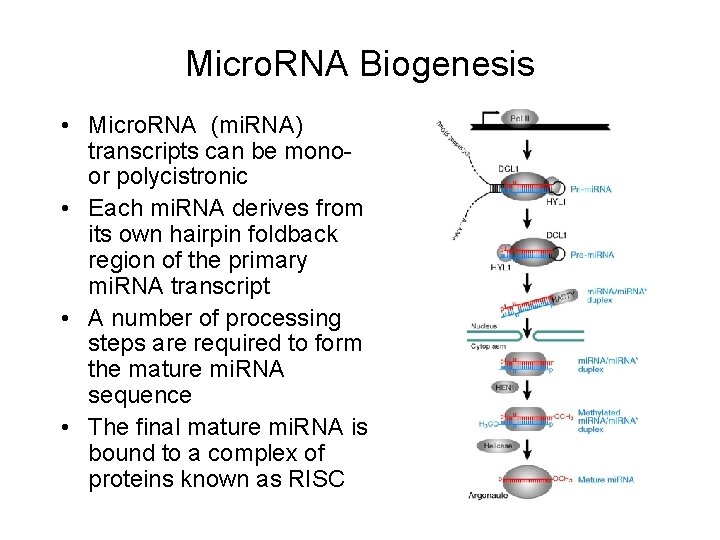
Micro. RNA Biogenesis • Micro. RNA (mi. RNA) transcripts can be monoor polycistronic • Each mi. RNA derives from its own hairpin foldback region of the primary mi. RNA transcript • A number of processing steps are required to form the mature mi. RNA sequence • The final mature mi. RNA is bound to a complex of proteins known as RISC

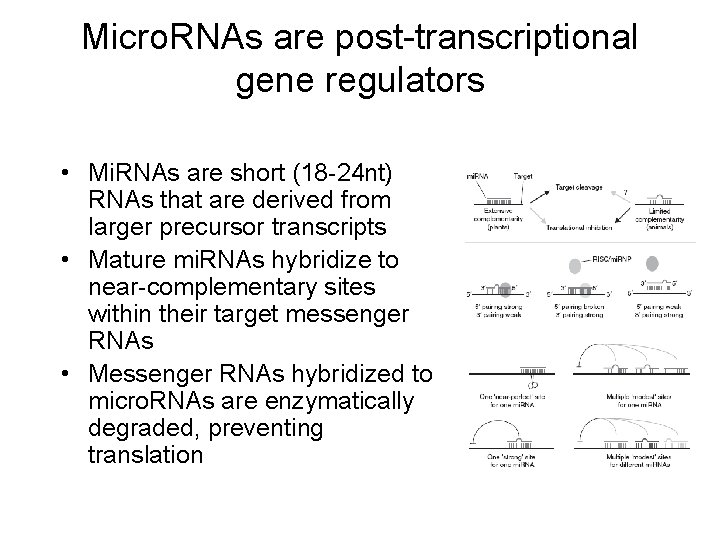
Micro. RNAs are post-transcriptional gene regulators • Mi. RNAs are short (18 -24 nt) RNAs that are derived from larger precursor transcripts • Mature mi. RNAs hybridize to near-complementary sites within their target messenger RNAs • Messenger RNAs hybridized to micro. RNAs are enzymatically degraded, preventing translation

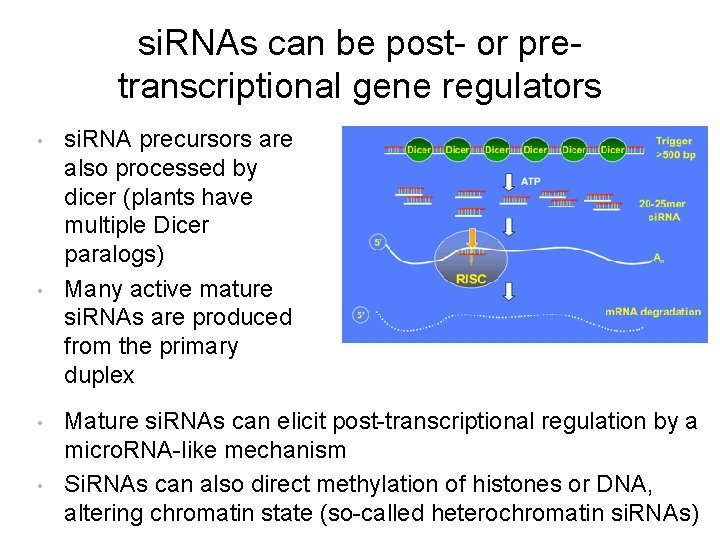
si. RNAs can be post- or pretranscriptional gene regulators • • si. RNA precursors are also processed by dicer (plants have multiple Dicer paralogs) Many active mature si. RNAs are produced from the primary duplex Mature si. RNAs can elicit post-transcriptional regulation by a micro. RNA-like mechanism Si. RNAs can also direct methylation of histones or DNA, altering chromatin state (so-called heterochromatin si. RNAs)

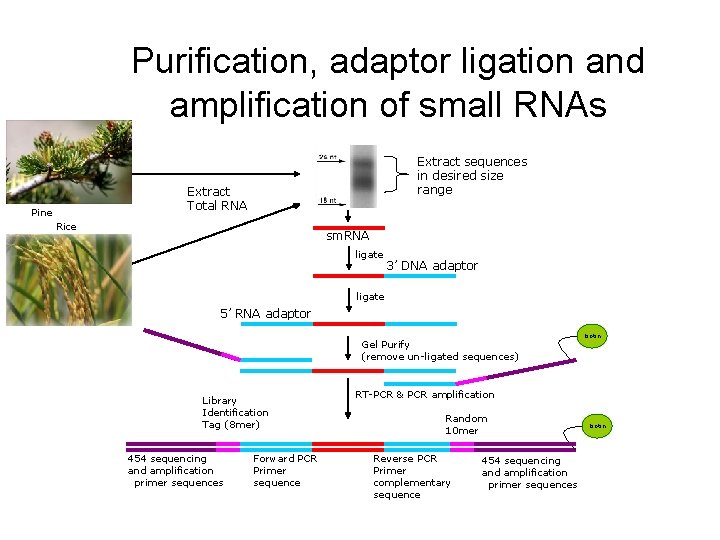
Purification, adaptor ligation and amplification of small RNAs Extract sequences in desired size range Extract Total RNA Pine Rice sm. RNA ligate 3’ DNA adaptor ligate 5’ RNA adaptor Gel Purify (remove un-ligated sequences) Library Identification Tag (8 mer) 454 sequencing and amplification primer sequences Forward PCR Primer sequence biotin RT-PCR & PCR amplification Random 10 mer Reverse PCR Primer complementary sequence 454 sequencing and amplification primer sequences biotin

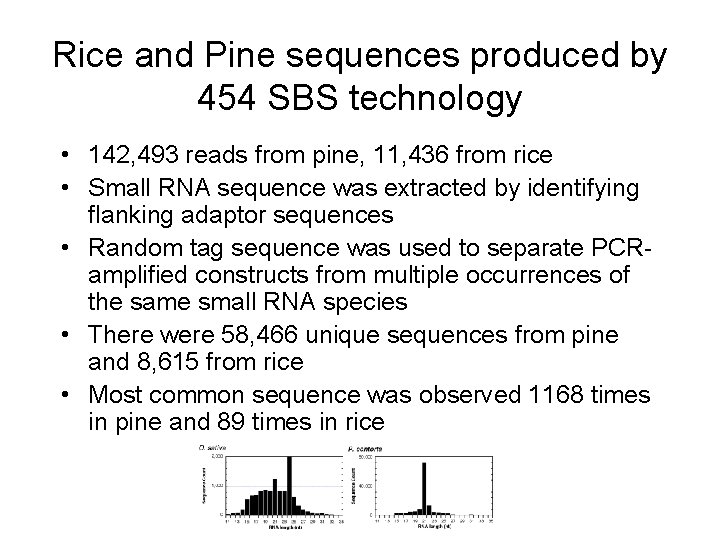
Rice and Pine sequences produced by 454 SBS technology • 142, 493 reads from pine, 11, 436 from rice • Small RNA sequence was extracted by identifying flanking adaptor sequences • Random tag sequence was used to separate PCRamplified constructs from multiple occurrences of the same small RNA species • There were 58, 466 unique sequences from pine and 8, 615 from rice • Most common sequence was observed 1168 times in pine and 89 times in rice

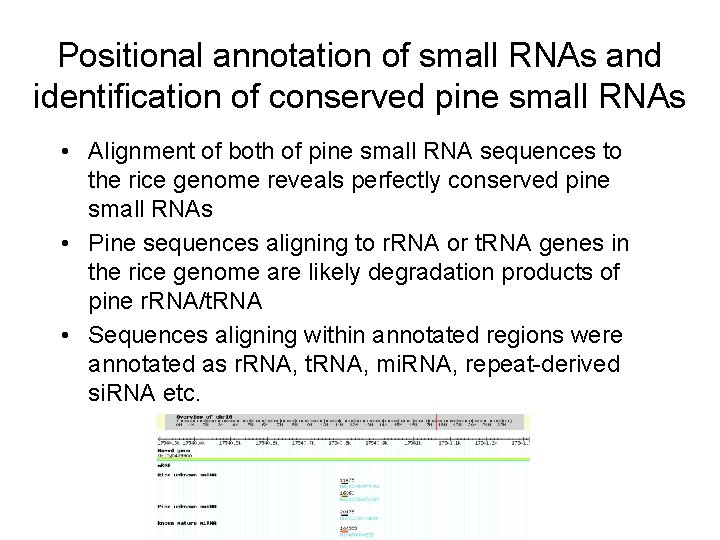
Positional annotation of small RNAs and identification of conserved pine small RNAs • Alignment of both of pine small RNA sequences to the rice genome reveals perfectly conserved pine small RNAs • Pine sequences aligning to r. RNA or t. RNA genes in the rice genome are likely degradation products of pine r. RNA/t. RNA • Sequences aligning within annotated regions were annotated as r. RNA, t. RNA, mi. RNA, repeat-derived si. RNA etc.

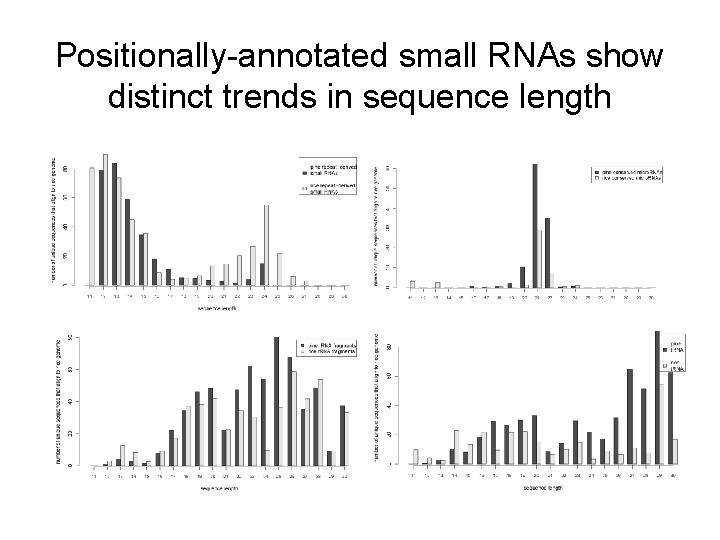
Positionally-annotated small RNAs show distinct trends in sequence length

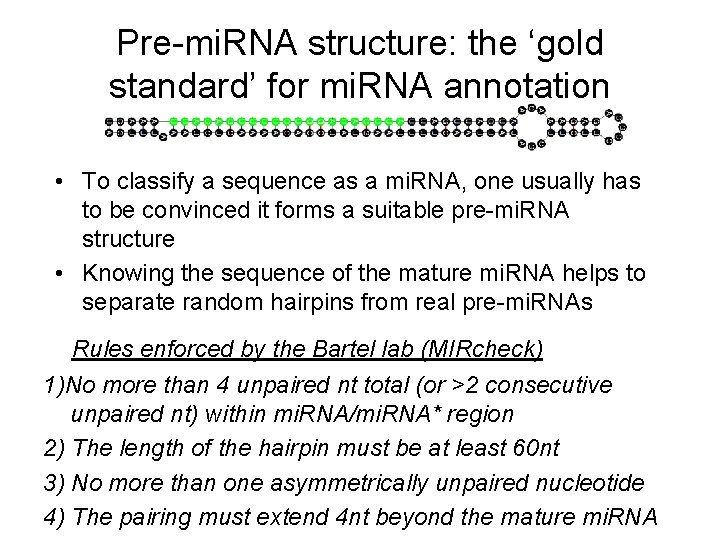
Pre-mi. RNA structure: the ‘gold standard’ for mi. RNA annotation • To classify a sequence as a mi. RNA, one usually has to be convinced it forms a suitable pre-mi. RNA structure • Knowing the sequence of the mature mi. RNA helps to separate random hairpins from real pre-mi. RNAs Rules enforced by the Bartel lab (MIRcheck) 1)No more than 4 unpaired nt total (or >2 consecutive unpaired nt) within mi. RNA/mi. RNA* region 2) The length of the hairpin must be at least 60 nt 3) No more than one asymmetrically unpaired nucleotide 4) The pairing must extend 4 nt beyond the mature mi. RNA

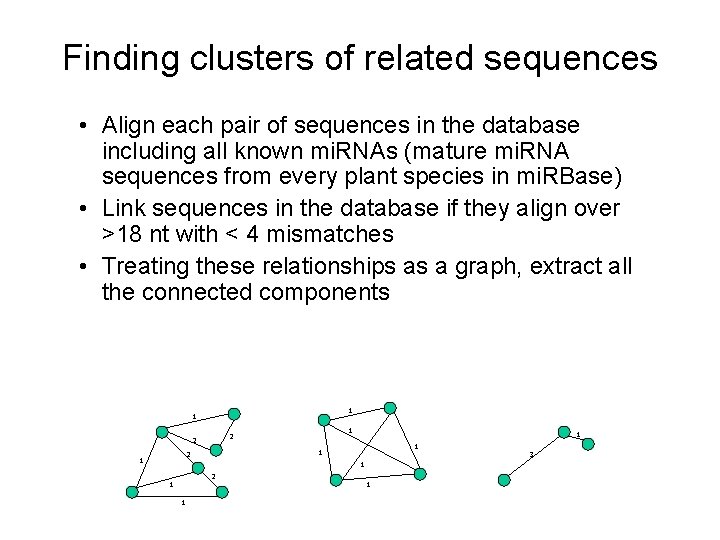
Finding clusters of related sequences • Align each pair of sequences in the database including all known mi. RNAs (mature mi. RNA sequences from every plant species in mi. RBase) • Link sequences in the database if they align over >18 nt with < 4 mismatches • Treating these relationships as a graph, extract all the connected components 1 1 1 2 2 1 1 1 3

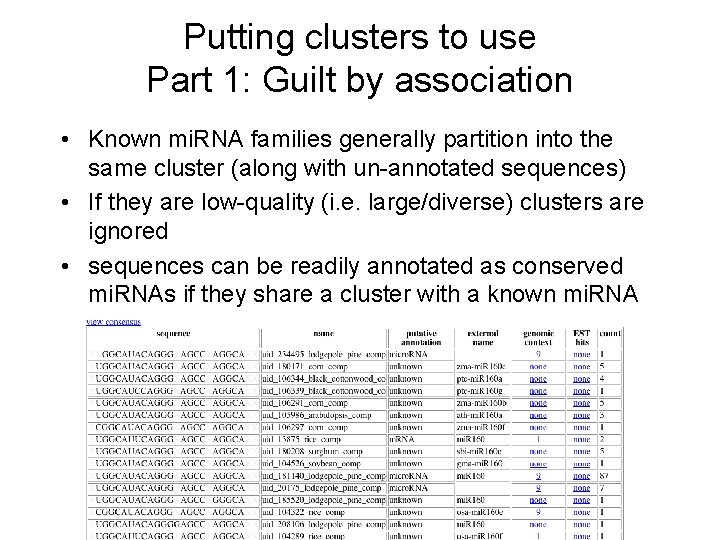
Putting clusters to use Part 1: Guilt by association • Known mi. RNA families generally partition into the same cluster (along with un-annotated sequences) • If they are low-quality (i. e. large/diverse) clusters are ignored • sequences can be readily annotated as conserved mi. RNAs if they share a cluster with a known mi. RNA

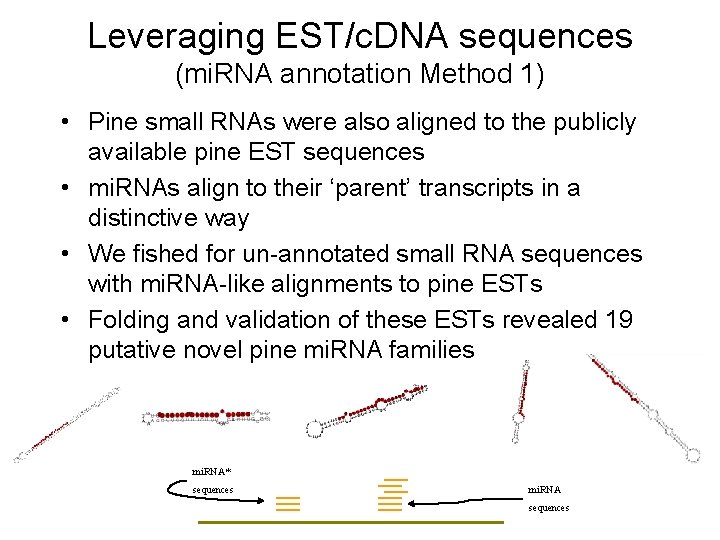
Leveraging EST/c. DNA sequences (mi. RNA annotation Method 1) • Pine small RNAs were also aligned to the publicly available pine EST sequences • mi. RNAs align to their ‘parent’ transcripts in a distinctive way • We fished for un-annotated small RNA sequences with mi. RNA-like alignments to pine ESTs • Folding and validation of these ESTs revealed 19 putative novel pine mi. RNA families mi. RNA* sequences mi. RNA sequences

Putting clusters to use part 2: mi. RNA-like Clusters (method 2) • Clusters of known mi. RNA families had many similarities which might allow their separation from non-mi. RNA clusters – – High information content Mean sequence length Low variance in sequence length Few indels, relatively few genomic loci • A support vector machine (SVM) classifier was trained on all known micro. RNA clusters • This classifier demonstrated high specificity (0. 96) and modest sensitivity (0. 65) • 54 clusters were predicted to be mi. RNA clusters (all pine-specific) and may represent novel pine micro. RNA families • 5 of these clusters are confirmed mi. RNAs based on EST folding

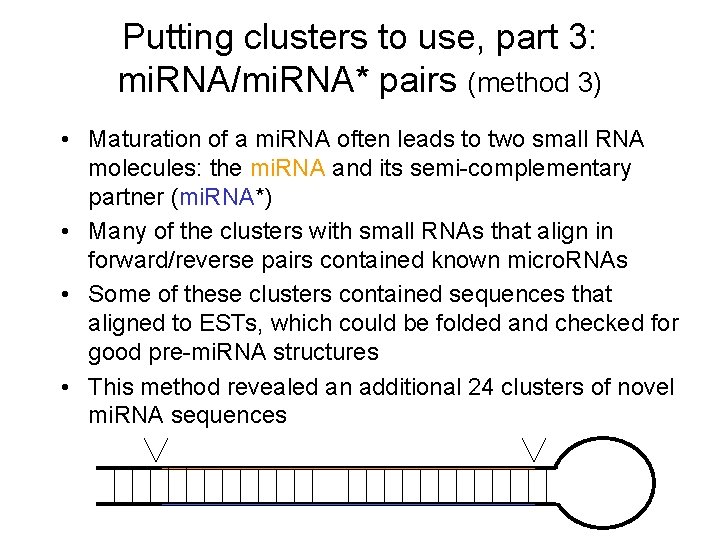
Putting clusters to use, part 3: mi. RNA/mi. RNA* pairs (method 3) • Maturation of a mi. RNA often leads to two small RNA molecules: the mi. RNA and its semi-complementary partner (mi. RNA*) • Many of the clusters with small RNAs that align in forward/reverse pairs contained known micro. RNAs • Some of these clusters contained sequences that aligned to ESTs, which could be folded and checked for good pre-mi. RNA structures • This method revealed an additional 24 clusters of novel mi. RNA sequences

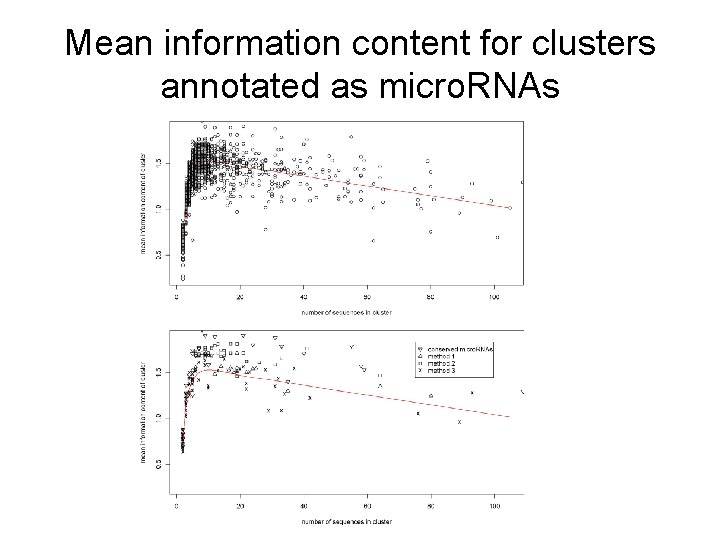
Mean information content for clusters annotated as micro. RNAs

Summary • Some of the 24 nt heterachromatin si. RNAs (including repeat-derived si. RNAs) are conserved between gymnosperms and angiosperms • Pine small RNA sequences are rich in conserved micro. RNAs • Various methods, including sequence clustering, RNA folding and machine learning, reveal many putative novel pine mi. RNA families

Acknowledgements Thanks: Peter Unrau (SFU, MBB) Cenk Sahinalp & his lab (SFU, CS) Gozde Cozen (SFU, CS) Alex Ebhardt (SFU, MBB) Elena Dolgosheina (SFU, MBB) Matt Hickenbotham (Wash. U) Vincent Magrini (Wash. U) The Van. BUG Dev team Other Van. BUG sponsors: IBM, Genome BC

Alignment of 3 ESTs from a novel pine mi. RNA family