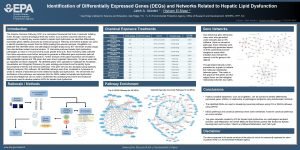
Identification and analysis of differentially expressed genes in







- Slides: 20

Identification and analysis of differentially expressed genes in Saccharomyces cerevisiae. Group Populus: Petra van Berkel Casper Gerritsen Astri Herlino Brian Lavrijssen

Dataset of S. cerevisiae �Data generated by Nookaew et al (2012) �Two conditions: �Glucose excess (Batch) & Glucose limited (Chemostat) � 3 Biological replicates per condition �RNA-seq data: � 12 Files � 3 Sets of Paired-end reads per condition �Pipeline for differential gene expression analysis

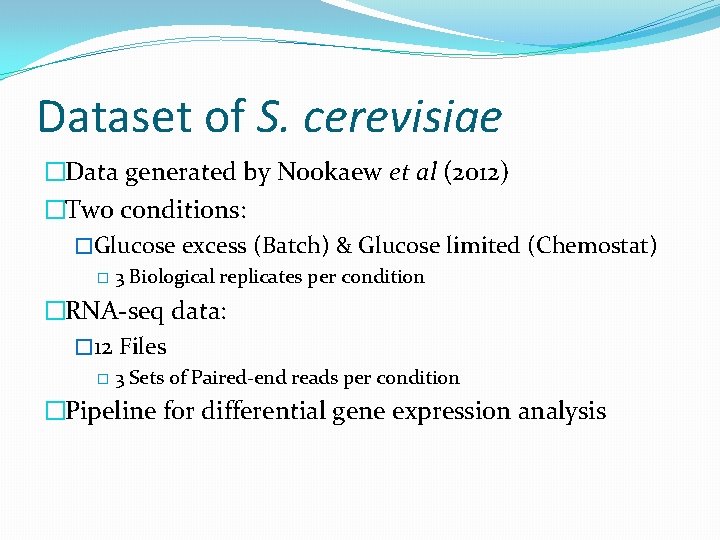
Top. Hat – Cufflinks analysis �Protocols based on Trapnell et al (2012) � 75% of reads mapped �Plots based on Cuffdiff gene expression output

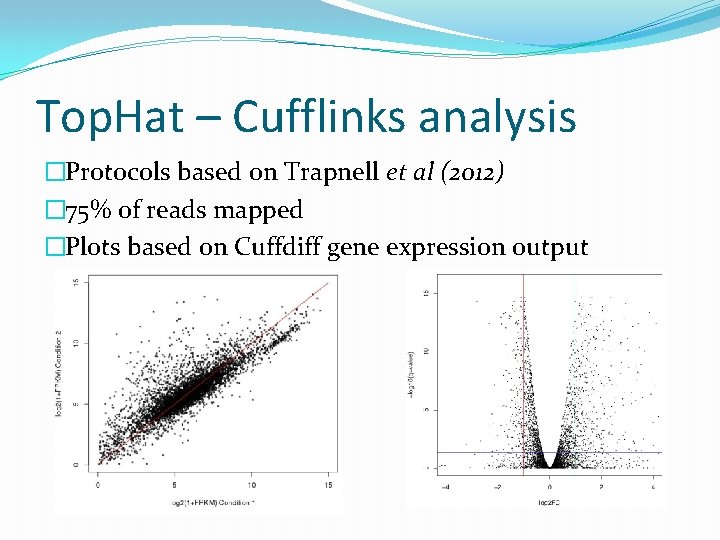
Cuffdiff output • 5800 genes with FPKM values • Q-value threshold based on Nookaew et al (2012) Data Summary Significant differentially expressed FPKM > 0 and value_2 > 0 log 2(fold change) > 1 log 2(fold change) < -1 log 2(fold change) > 3 log 2(fold change) > -2 Q-value < 0. 05 2560 2554 735 510 177 44 Q-value < 1 e-5 1293 1292 516 410 151 33

Validation of Top. Hat - Cufflinks �Validation of selection �Using Excel �Literature study �Boer et al (2003) � Influence of C, N, P and S limitation � Microarray analysis � > 68 out of 151 significantly upregulated � > 9 out of 33 significantly downregulated �More or less same genes found in other papers

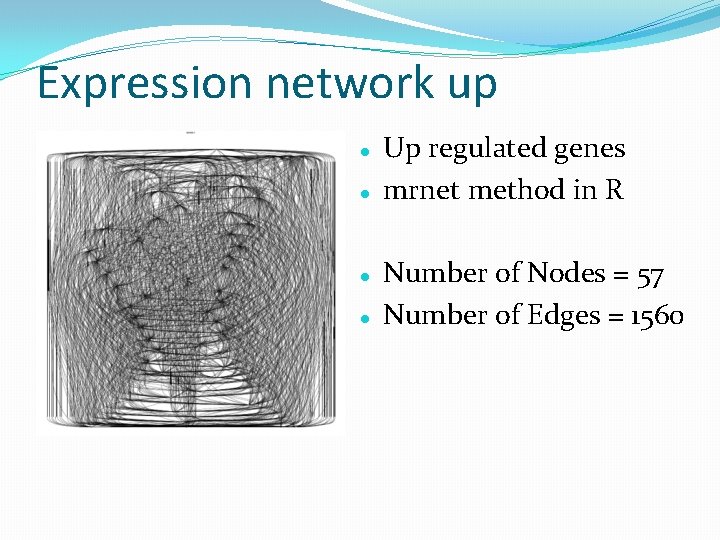
Expression network up Up regulated genes mrnet method in R Number of Nodes = 57 Number of Edges = 1560

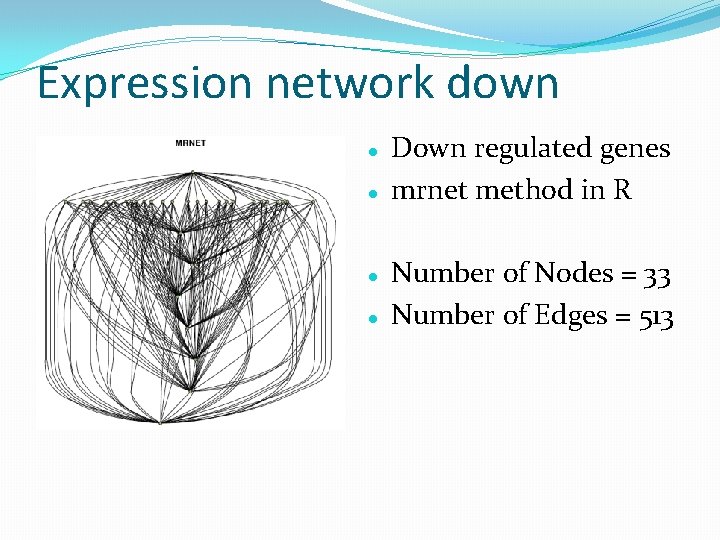
Expression network down Down regulated genes mrnet method in R Number of Nodes = 33 Number of Edges = 513

GO Terms and GO Enrichment R version 2. 15. 0 (2012 -03 -30) �Packages: �bioma. Rt: Ensembl gene 69, S. cerevisiae EF 3 �org. Sc. sgd. db �GOstats �Rgraphviz �GO enrichment: � 8419 genes in the universe (org. Sc. sgd. PMID 2 ORF) �Threshold: p-value < 10 -4

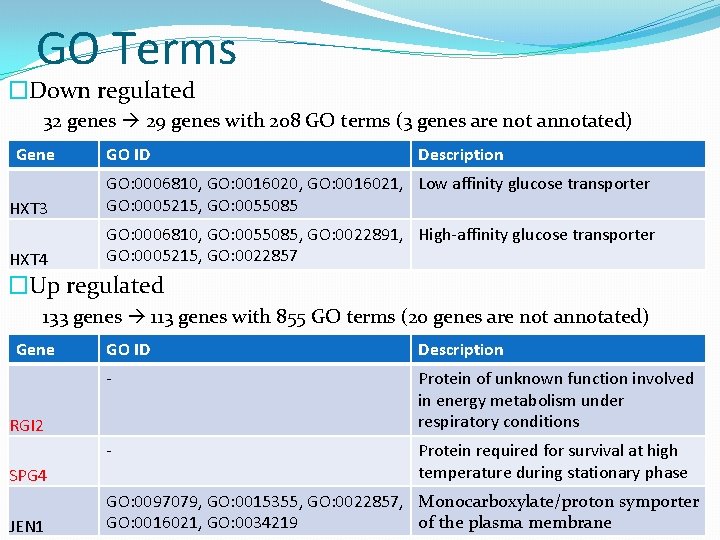
GO Terms �Down regulated 32 genes 29 genes with 208 GO terms (3 genes are not annotated) Gene GO ID Description HXT 3 GO: 0006810, GO: 0016021, Low affinity glucose transporter GO: 0005215, GO: 0055085 HXT 4 GO: 0006810, GO: 0055085, GO: 0022891, High-affinity glucose transporter GO: 0005215, GO: 0022857 �Up regulated 133 genes 113 genes with 855 GO terms (20 genes are not annotated) Gene GO ID Description - Protein of unknown function involved in energy metabolism under respiratory conditions - Protein required for survival at high temperature during stationary phase RGI 2 SPG 4 JEN 1 GO: 0097079, GO: 0015355, GO: 0022857, Monocarboxylate/proton symporter GO: 0016021, GO: 0034219 of the plasma membrane

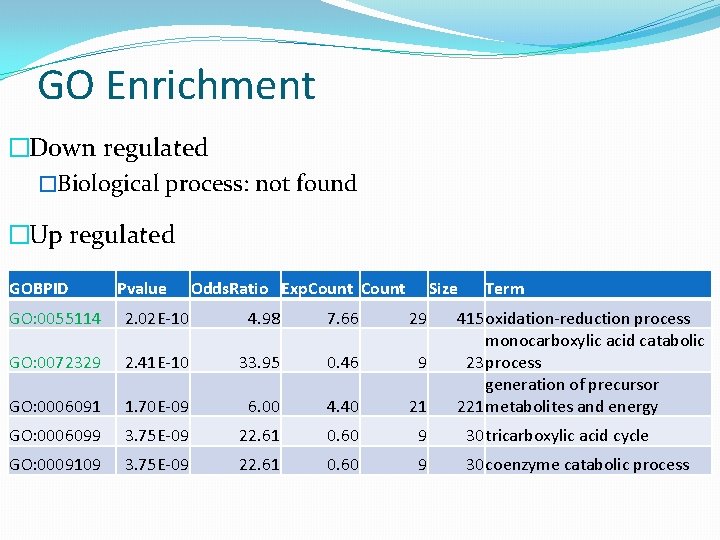
GO Enrichment �Down regulated �Biological process: not found �Up regulated GOBPID Pvalue Odds. Ratio Exp. Count Size Term GO: 0055114 2. 02 E-10 4. 98 7. 66 29 415 oxidation-reduction process monocarboxylic acid catabolic 23 process generation of precursor 221 metabolites and energy GO: 0072329 2. 41 E-10 33. 95 0. 46 9 GO: 0006091 1. 70 E-09 6. 00 4. 40 21 GO: 0006099 3. 75 E-09 22. 61 0. 60 9 30 tricarboxylic acid cycle GO: 0009109 3. 75 E-09 22. 61 0. 60 9 30 coenzyme catabolic process

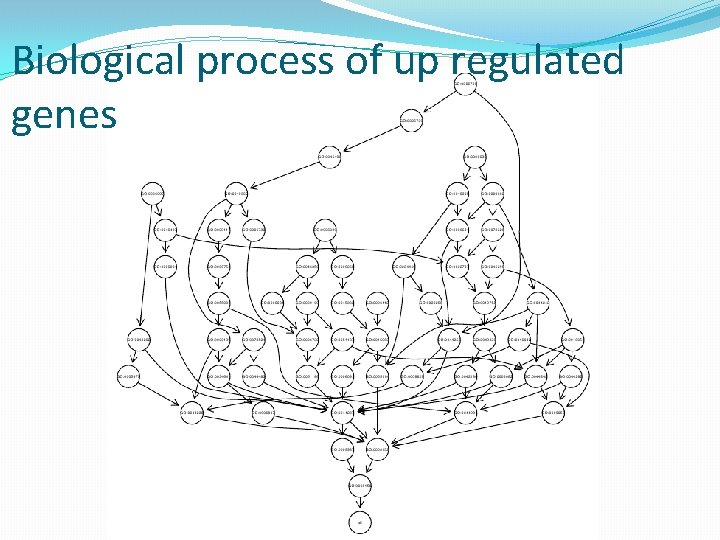
Biological process of up regulated genes

�Validation: Yeast genome database �Problem: �Not well annotated because the bioma. Rt was not updated to Ensembl gene 70, S. cerevisiae EF 4

Top 100 �gffread: make the transcripts fasta file �Determine the top 100 highest and lowest expressed genes for the two conditions �R: order cuffdiff output on FPKM value (4 files) �Take out the genes with FPKM = 0

Top 100 �Top genes: �G 3 P dehydrogenase, �F 16 P aldolase, �Ribosomal subunit protein �Bottom genes: �dubious transcript, �retro transposon, �etc. .

GC-content & transcript length �Determine GC-content and transcript length �Import top 100 genes files �For each file check the genes in top 100 file in transcripts. fa and count GC content and the transcript length

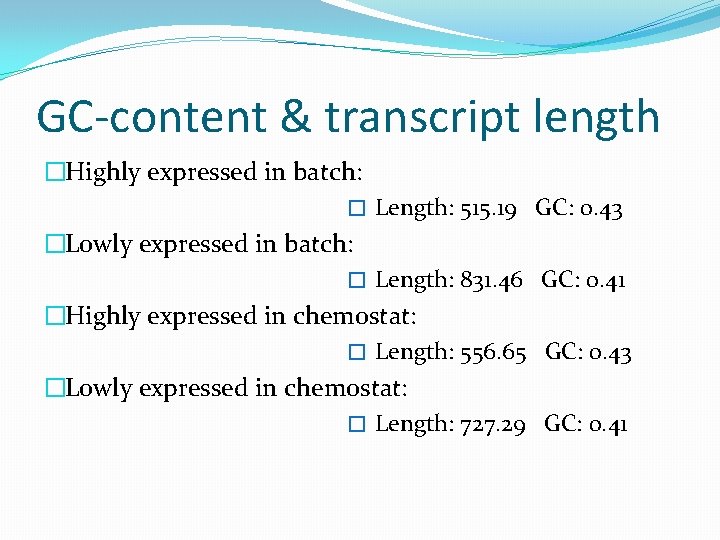
GC-content & transcript length �Highly expressed in batch: � Length: 515. 19 GC: 0. 43 �Lowly expressed in batch: � Length: 831. 46 GC: 0. 41 �Highly expressed in chemostat: � Length: 556. 65 GC: 0. 43 �Lowly expressed in chemostat: � Length: 727. 29 GC: 0. 41

GC-content & transcript length �Short sequence length! �mainly in highly expressed genes, gives unrealistic view of codon usage and intron length �These are often ribosomal subunit proteins

Intron length Genes. gtf as input Create an indexfile Look for the interesting genes Print them to an outputfile Calculate average file mean intron length introns_hi 1. out 429. 455 introns_hi 2. out 440. 125 introns_low 1. out 60. 6667 introns_low 2. out 43. 5

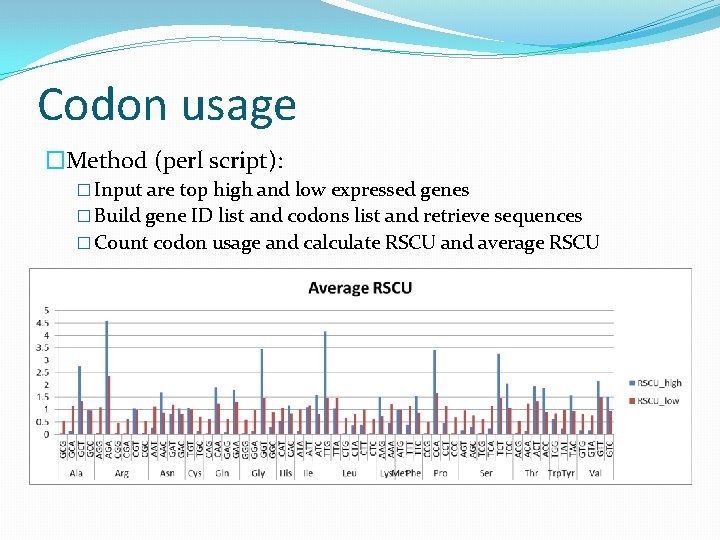
Codon usage �Method (perl script): � Input are top high and low expressed genes � Build gene ID list and codons list and retrieve sequences � Count codon usage and calculate RSCU and average RSCU

Conclusion �The up and down regulated genes are involved in carbon metabolism �Highly expressed genes are involved in carbon metabolism or are ribosomal subunit proteins
 Linked genes and unlinked genes
Linked genes and unlinked genes Linked genes and unlinked genes
Linked genes and unlinked genes Differentially
Differentially Factors of 15
Factors of 15 Central pocket whorl vs plain whorl
Central pocket whorl vs plain whorl Scanning and analysis tools are used to pinpoint
Scanning and analysis tools are used to pinpoint Hemizygous definition biology
Hemizygous definition biology Stabilizing selection human birth weight
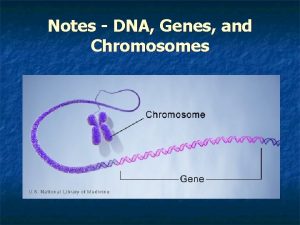
Stabilizing selection human birth weight The relationship between genes dna and chromosomes
The relationship between genes dna and chromosomes How do you know if a karyotype is male or female
How do you know if a karyotype is male or female Genes is the study of heredity and variation
Genes is the study of heredity and variation This section describes
This section describes Chapter 11 dna and genes
Chapter 11 dna and genes Evolution of populations section 16-1 genes and variation
Evolution of populations section 16-1 genes and variation Chapter 16 evolution of populations
Chapter 16 evolution of populations Dominant and recessive genes
Dominant and recessive genes Chromosomes genes and basic genetics foldable answer key
Chromosomes genes and basic genetics foldable answer key Dna, genes and chromosomes relationship
Dna, genes and chromosomes relationship Dna and genes chapter 11
Dna and genes chapter 11 Section 16-1 genes and variation
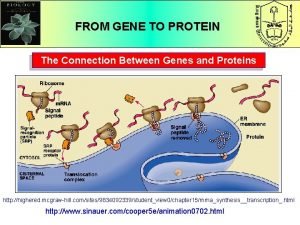
Section 16-1 genes and variation What is the connection between genes and proteins
What is the connection between genes and proteins