Detection of Significantly Differentially Expressed Genes Statistical tests












- Slides: 39

Detection of Significantly Differentially Expressed Genes Statistical tests • Student’s t test for two conditions/groups (control vs treated) (i. e. the comparison of the means and standard deviations of two bell shaped curves, based on a t-statistic, testing the nullhypothesis that both distributions came from the same distribution) • ANOVA analysis (control vs treatment 1 vs treatment 2) (i. e. ANalysis Of VAriances: Allows to test the null hypothesis that the differences within and between at least 3 groups are the same on average. Based on F-statistic, the ratio of the variance calculated among the means to the variance within the samples)

Detection of Significantly Differentially Expressed Genes • 2 -way ANOVA (eg 2 cell lines, 2 treatments) (i. e. The two-way ANOVA compares the mean differences between groups that have been split on two independent variables called factors. The primary purpose of a two-way ANOVA is to understand if there is an interaction between the two independent variables on the dependent variable). • SAM analysis (Signifiance Analysis of Microarrays) Significant genes are determined through permutation tests.

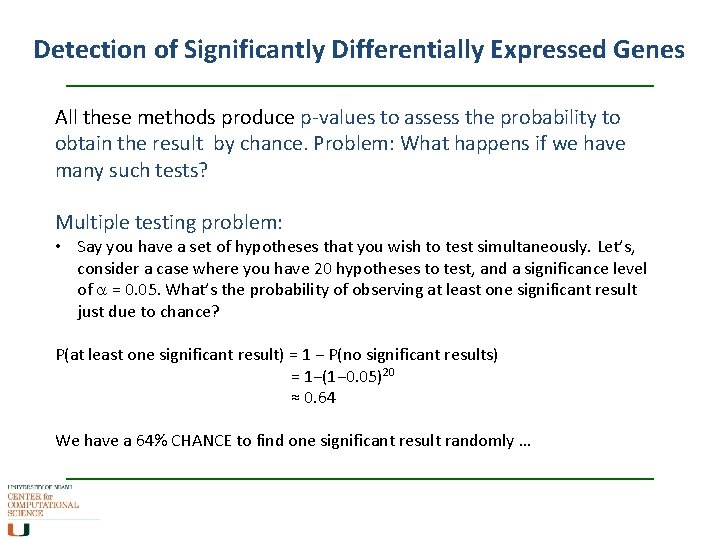
Detection of Significantly Differentially Expressed Genes All these methods produce p-values to assess the probability to obtain the result by chance. Problem: What happens if we have many such tests? Multiple testing problem: • Say you have a set of hypotheses that you wish to test simultaneously. Let’s, consider a case where you have 20 hypotheses to test, and a significance level of a = 0. 05. What’s the probability of observing at least one significant result just due to chance? P(at least one significant result) = 1 − P(no significant results) = 1−(1− 0. 05)20 ≈ 0. 64 We have a 64% CHANCE to find one significant result randomly …

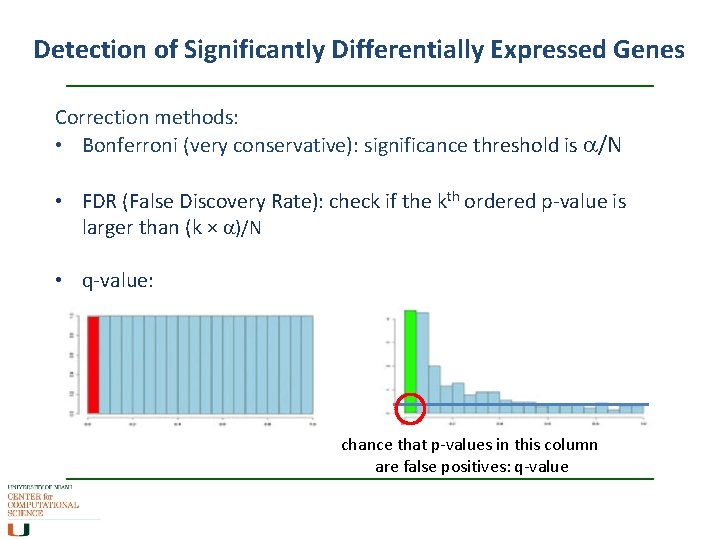
Detection of Significantly Differentially Expressed Genes Correction methods: • Bonferroni (very conservative): significance threshold is a/N • FDR (False Discovery Rate): check if the kth ordered p-value is larger than (k × a)/N • q-value: chance that p-values in this column are false positives: q-value

Detection of Significantly Differentially Expressed Genes Fold change • Difference in the intensity of a sample vs control or another sample, indicative of difference in level of expression of the gene • Threshold > 2, or > 1. 6 in some cases

Then clustering

Then clustering • In differential gene expression, you are looking for genes that behave differently between one sample and another, either up- or down • Once you get your DE gene set, you group the genes according to similar expression, and the outliers become more obvious • Clustering methods similar to those of phylogenetics, but without the evolutionary weightings, ie distance matrices More downstream analysis later in the course

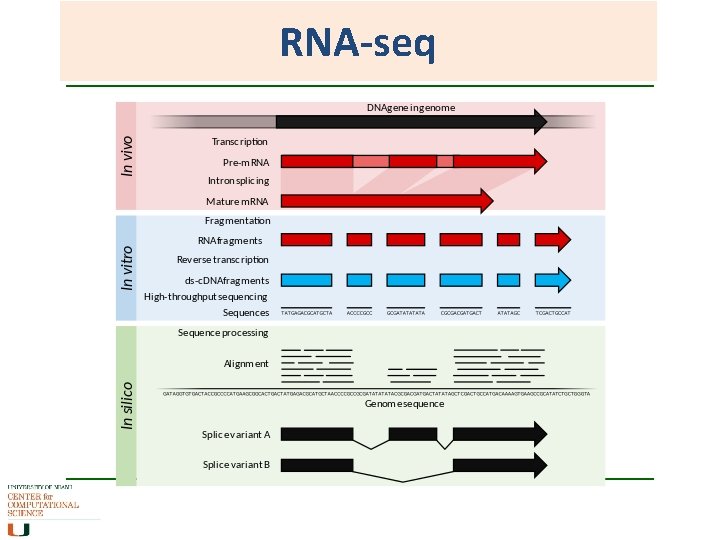
RNA-seq Same concept as sequencing ESTs and counting SAGE tags, but does not stop at short segments and tags. What is being sequenced is the c. DNA from the m. RNA component. Sequencing of whole transcriptome of a sample (NGS), and comparing it against the whole transcriptome of another sample. Costly, informative, bioinformatics not yet fully sorted outwhen does a lot of data become too much data?

RNA-seq

Finding the real transcripts

It’s all about the alignment • First, you align your reads to a reference genome or genomic region (or assemble the reads de novo) BWA, Bowtie 2, etc • Then you use a splice-aware aligner, such as Top. Hat or STAR, to refine the aligments according to coding sequences (exons) using known and/or predicted splice junctions

Quantifying reads per gene • Your aim is to count sequence reads per gene • When mapping reads to genome: Filter out r. RNA, t. RNA, mit. RNA, etc Filtering out (or in!) non-coding RNA Deal with alternative splicing Deal with overlapping genes, pseudogenes Small reads mean many short overlaps at one end or the other of intron gaps • Allele specific gene expression • • •

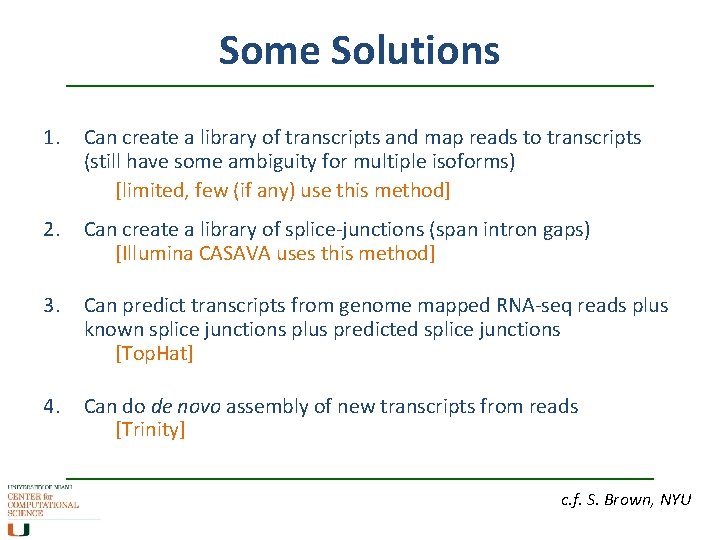
Some Solutions 1. Can create a library of transcripts and map reads to transcripts (still have some ambiguity for multiple isoforms) [limited, few (if any) use this method] 2. Can create a library of splice-junctions (span intron gaps) [Illumina CASAVA uses this method] 3. Can predict transcripts from genome mapped RNA-seq reads plus known splice junctions plus predicted splice junctions [Top. Hat] 4. Can do de novo assembly of new transcripts from reads [Trinity] c. f. S. Brown, NYU

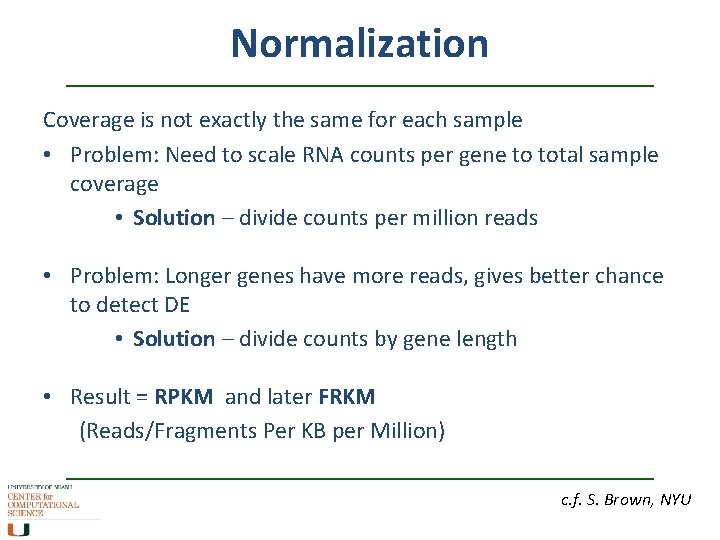
Normalization Coverage is not exactly the same for each sample • Problem: Need to scale RNA counts per gene to total sample coverage • Solution – divide counts per million reads • Problem: Longer genes have more reads, gives better chance to detect DE • Solution – divide counts by gene length • Result = RPKM and later FRKM (Reads/Fragments Per KB per Million) c. f. S. Brown, NYU

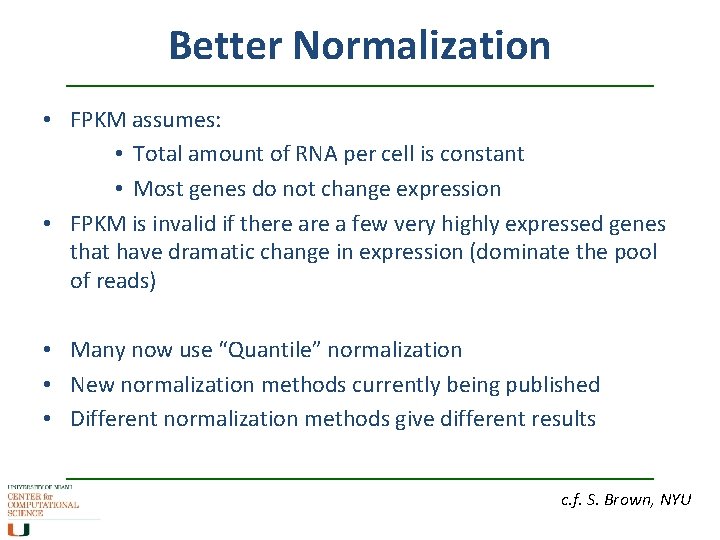
Better Normalization • FPKM assumes: • Total amount of RNA per cell is constant • Most genes do not change expression • FPKM is invalid if there a few very highly expressed genes that have dramatic change in expression (dominate the pool of reads) • Many now use “Quantile” normalization • New normalization methods currently being published • Different normalization methods give different results c. f. S. Brown, NYU

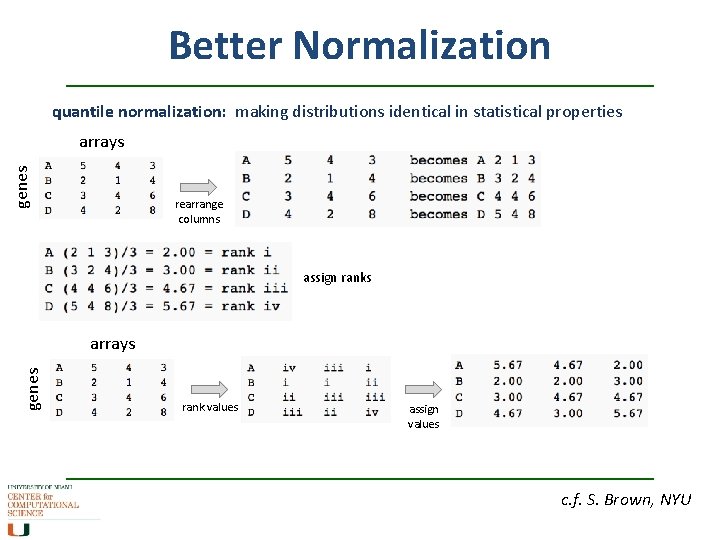
Better Normalization quantile normalization: making distributions identical in statistical properties genes arrays rearrange columns assign ranks genes arrays rank values assign values c. f. S. Brown, NYU

Statistics of Differential Gene Expression • m. RNA levels are variable in cells/tissues/organisms over time/treatment/tissue etc. • Need enough replicates to separate biological variability from experimental variability • If there is high experimental variability, then variance within replicates will be high, statistical significance for DE will be difficult to find. • Best methods to discover DE are coupled with sophisticated approaches to normalization • Very low expressing genes are tricky: FPKM<1 c. f. S. Brown, NYU

Gene Expression Analysis Databases: GEO from NCBI Array. Express from EBI Commercial software: Gene. Spring GX, CLC Bio, many others Free: Mostly R based Not being scared of statistics is an advantage New methods and algorithms continually being published Routine experiments are routine, innovative methods more care The really tricky part is the interpretation of the results

https: //github. com/ccsstudentmentors/tutorials/wiki/CCS-Student-Mentors---Tutorials

Suggested additional reading:

Biological Pathways A series of actions that lead to a certain product or change in the cell. Within a pathway, proteins can be activated, deactivated to various degrees, or completely inactivated. e. g. metabolioc, signalling, gene regulation pathways Reasonably well defined, knowledge on mechanisms incomplete, but it exists. Databases available.

Main Types of Pathways Metabolic pathways (biochemistry) A series of chemical reactions that converts one type of chemical to another. eg glycolysis converts glucose to pyruvate Signaling pathways (molecular biology) A series of binding events, sometimes involving chemical actions (eg phosphorylation), that relay a message. eg MAP-kinase cascade Gene regulation networks (molecular biology/genetics) A series of binding and chemical events that regulate gene expression. eg transcription factor binding, histone deacetylation

Databases abound • Most trusted and most commonly used is KEGG • Also well cited is the Reactome database • Bio. Carta has nice diagrams • There are others; some are species or disease specific (Pathguide has a list, may be slightly outdated)







Biological Networks A network forms when proteins from several pathways work together. Networks link many pathways into a model. e. g. A change in signaling leads to gene regulation which leads to a new (or more or less or somehow different) metabolic product. Delving into the unknown, data driven, “given me the results of these different analyses and let me see what comes up”.

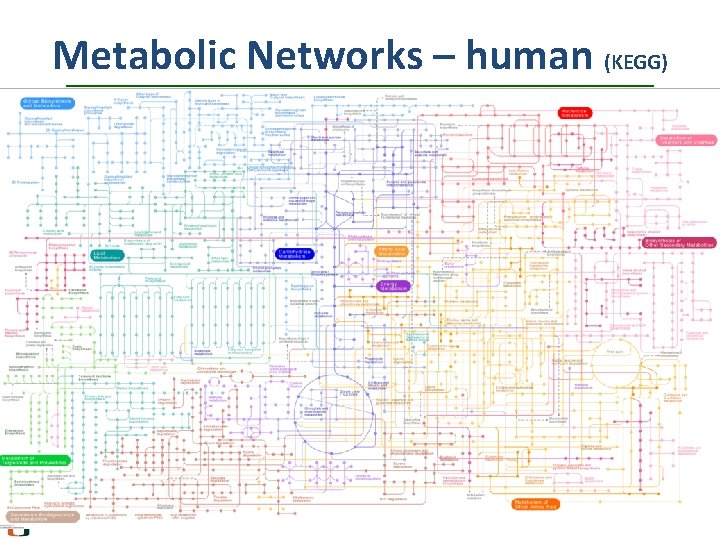
Metabolic Networks – human (KEGG)

Networks • Most work has been in protein-protein interactions. • Inferring biological networks, connecting the dots from pathway to pathway, or uncovering links previously not found between protein subsets. • Starting with pathway analysis of gene expression…

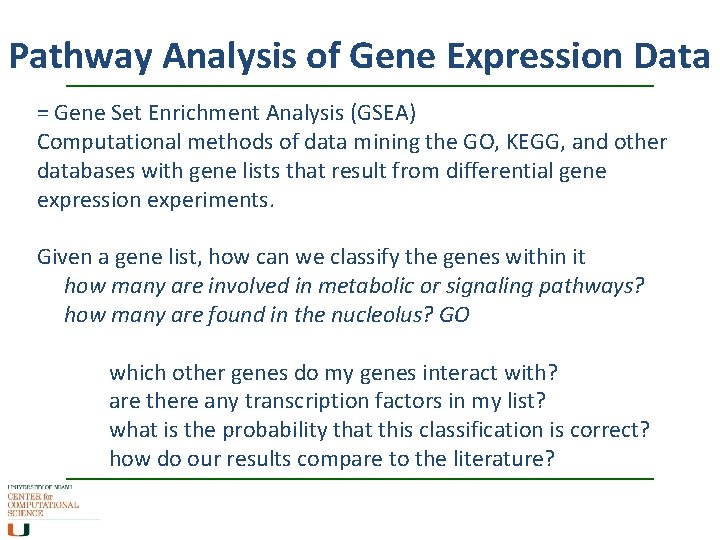
Pathway Analysis of Gene Expression Data = Gene Set Enrichment Analysis (GSEA) Computational methods of data mining the GO, KEGG, and other databases with gene lists that result from differential gene expression experiments. Given a gene list, how can we classify the genes within it how many are involved in metabolic or signaling pathways? how many are found in the nucleolus? GO which other genes do my genes interact with? are there any transcription factors in my list? what is the probability that this classification is correct? how do our results compare to the literature?

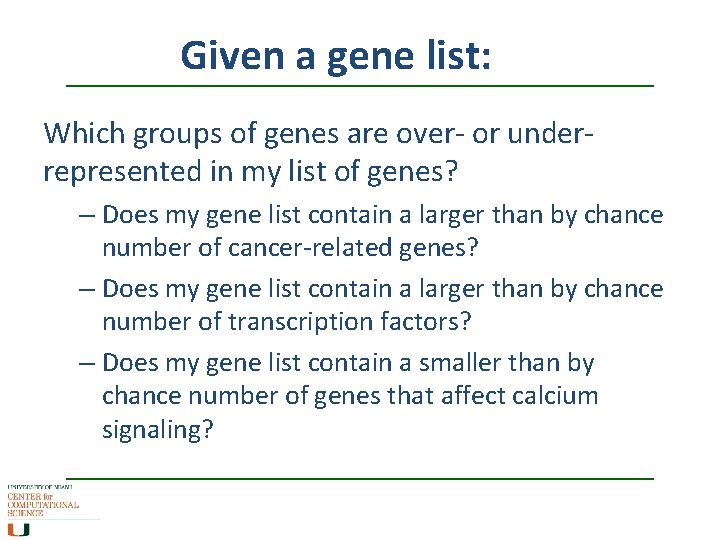
Given a gene list: Which groups of genes are over- or underrepresented in my list of genes? – Does my gene list contain a larger than by chance number of cancer-related genes? – Does my gene list contain a larger than by chance number of transcription factors? – Does my gene list contain a smaller than by chance number of genes that affect calcium signaling?

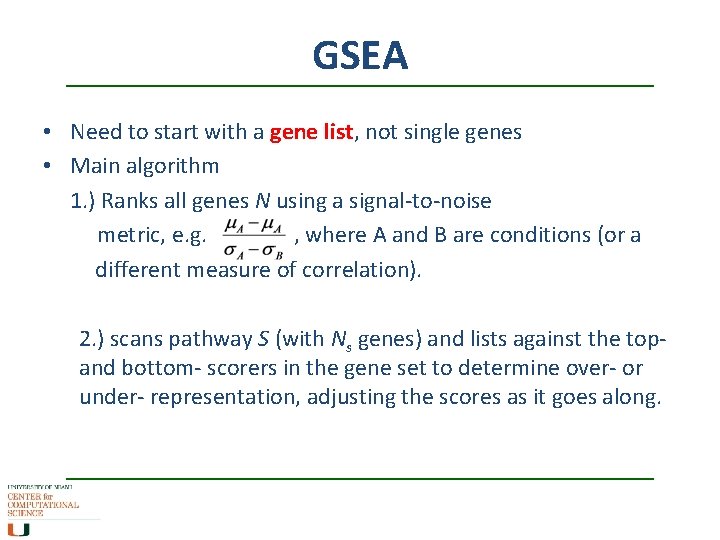
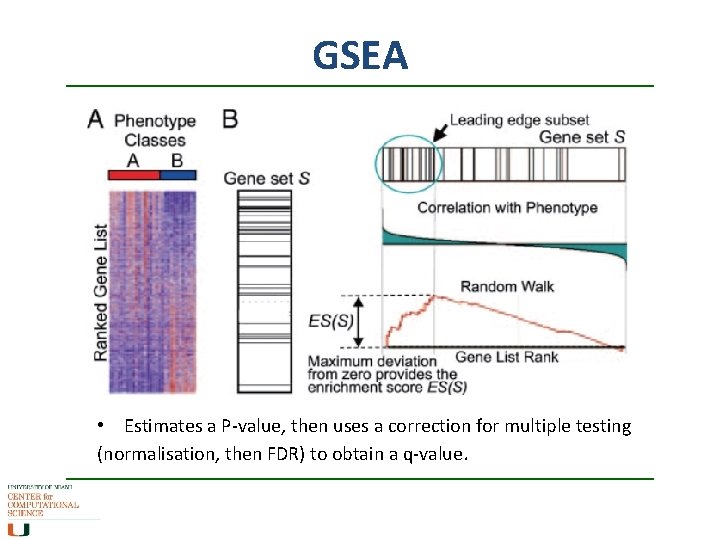
GSEA • Need to start with a gene list, not single genes • Main algorithm 1. ) Ranks all genes N using a signal-to-noise metric, e. g. , where A and B are conditions (or a different measure of correlation). 2. ) scans pathway S (with Ns genes) and lists against the topand bottom- scorers in the gene set to determine over- or under- representation, adjusting the scores as it goes along.

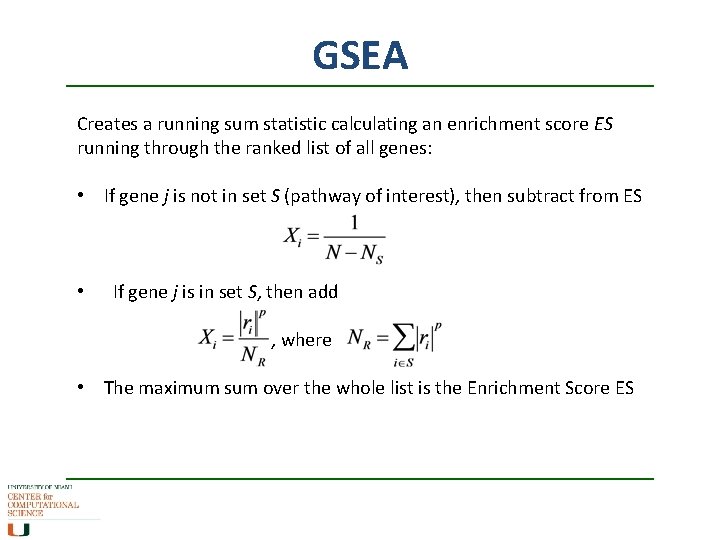
GSEA Creates a running sum statistic calculating an enrichment score ES running through the ranked list of all genes: • If gene j is not in set S (pathway of interest), then subtract from ES • If gene j is in set S, then add , where • The maximum sum over the whole list is the Enrichment Score ES

GSEA • Estimates a P-value, then uses a correction for multiple testing (normalisation, then FDR) to obtain a q-value.

Free Tools for Pathway Analysis There are “hundreds” of tools and algorithms out there For complicated investigations, may need a specialist to go through the options and work out a process best suited to your data. • • • GSEA-P (Subramanian, 2005, http: //www. ncbi. nlm. nih. gov/pubmed/17644558) Pathway Commons G: profiler DAVID Reactome • Gen. Sensor Suite (microarray only) • Flink (NCBI)

Licensed Software for Pathway Analysis Inexpensive Tools Pathway Studio from Elsevier Pathway analysis and literature text mining algorithm More Expensive Tools Meta. Core from Gene. Go metabolic, signaling and gene regulation maps; can integrate your own data IPA from Ingenuity