Hexaploid wheat Triticum aestivum 2 n 6 x



- Slides: 20

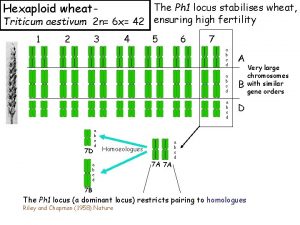
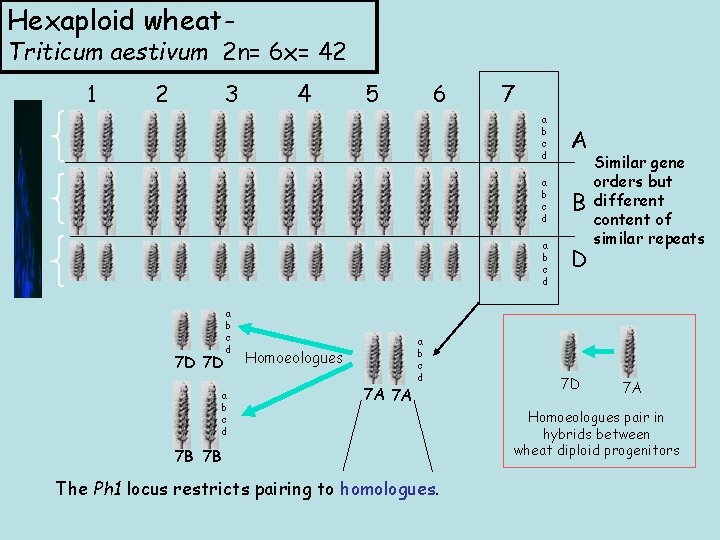
Hexaploid wheat- Triticum aestivum 2 n= 6 x= 42 1 2 3 4 5 6 7 a b c d A a b c d B a b c d 7 D 7 D a b c d Homoeologues 7 A 7 A a b c d 7 B 7 B The Ph 1 locus restricts pairing to homologues. D 7 D Similar gene orders but different content of similar repeats 7 A Homoeologues pair in hybrids between wheat diploid progenitors

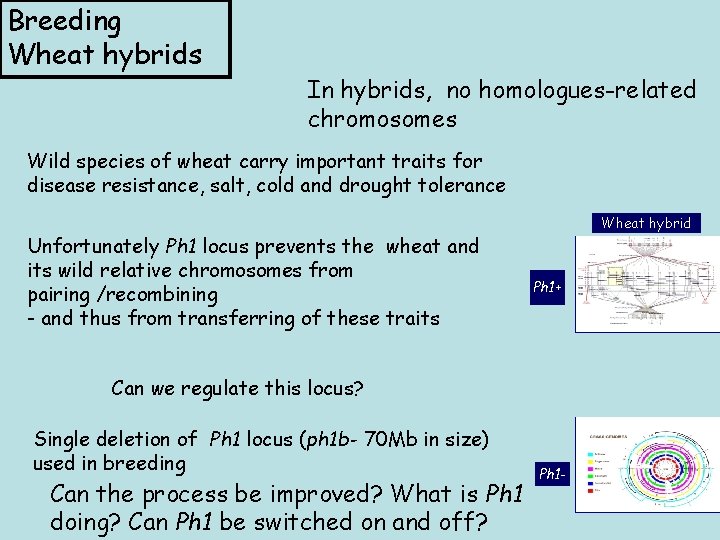
Breeding Wheat hybrids In hybrids, no homologues-related chromosomes Wild species of wheat carry important traits for disease resistance, salt, cold and drought tolerance Unfortunately Ph 1 locus prevents the wheat and its wild relative chromosomes from pairing /recombining - and thus from transferring of these traits Wheat hybrid Ph 1+ Can we regulate this locus? Single deletion of Ph 1 locus (ph 1 b- 70 Mb in size) used in breeding Can the process be improved? What is Ph 1 doing? Can Ph 1 be switched on and off? Ph 1 -

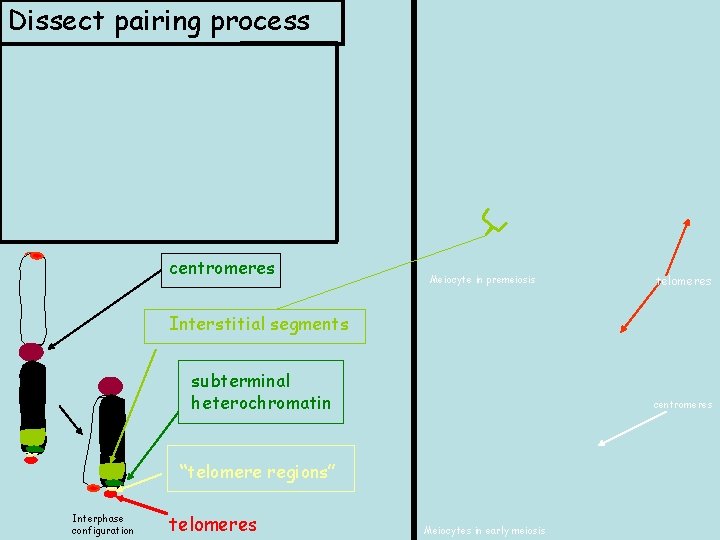
Dissect pairing process centromeres Meiocyte in premeiosis telomeres Interstitial segments subterminal heterochromatin centromeres “telomere regions” Interphase configuration telomeres Meiocytes in early meiosis

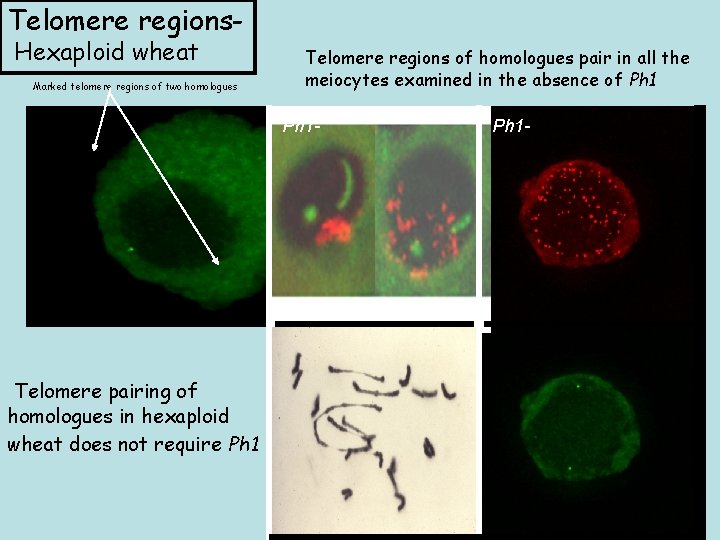
Telomere regions. Hexaploid wheat Marked telomere regions of two homologues Telomere regions of homologues pair in all the meiocytes examined in the absence of Ph 1 - Telomere pairing of homologues in hexaploid wheat does not require Ph 1 -

Summary of related issues • Kihara and Lilienfeld (1934) showed that wheat hybrids were prone to premature condensation- Implication- newly synthesized polyploids are prone to premature condensation • Premature/asynchronous condensation is a hallmark of cells that have an inadequate replication checkpoint and have begun condensation before completing replication • Overexpression of Cdc 2 (checkpoint) gene activity caused premature asynchronous condensation • Implication- newly synthesized polyploids need to control Cdc 2 activity- so what is Ph 1 affecting?

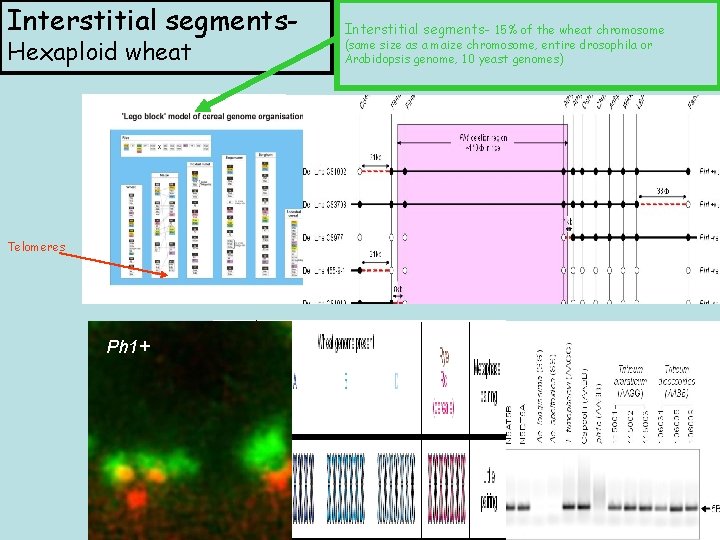
Interstitial segments. Hexaploid wheat Interstitial segments- 15% of the wheat chromosome (same size as a maize chromosome, entire drosophila or Arabidopsis genome, 10 yeast genomes) Ph 1 - Telomeres Ph 1+

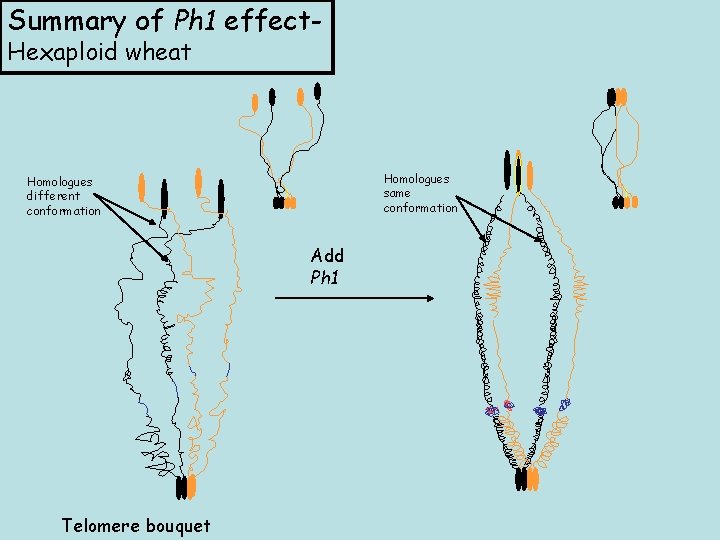
Summary of Ph 1 effect. Hexaploid wheat Homologues same conformation Homologues different conformation Add Ph 1 Telomere bouquet

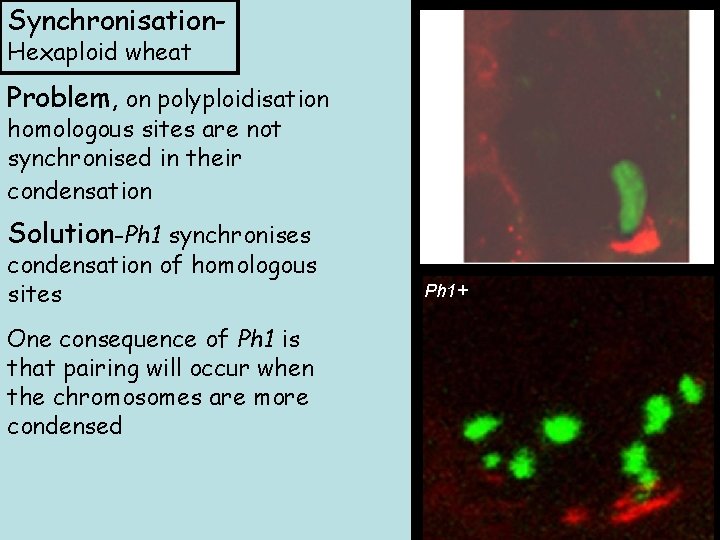
Synchronisation. Hexaploid wheat Ph 1 - Problem, on polyploidisation homologous sites are not synchronised in their condensation Solution-Ph 1 synchronises condensation of homologous sites One consequence of Ph 1 is that pairing will occur when the chromosomes are more condensed Ph 1+

Wheat hybrids Ph 1’s effect? In hybrids, no homologues-related chromosomes Heterochromatin (B chromosomes) can compensate for the absence of Ph 1 in wheat hybridsie suppress pairing and recombination between wheat and wild relative chromosomes Heterochromatin (B chromosomes) delay S phase and cause greater compaction of chromosome regions

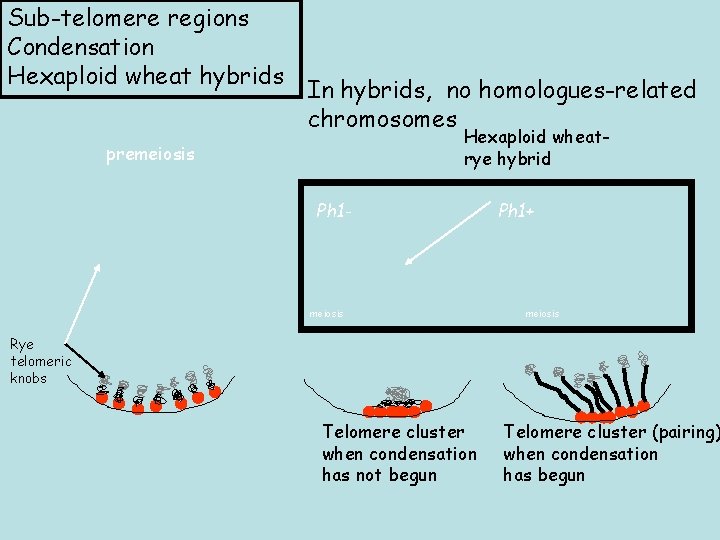
Sub-telomere regions Condensation Hexaploid wheat hybrids In hybrids, no homologues-related chromosomes Hexaploid wheatrye hybrid premeiosis Ph 1 - meiosis Ph 1+ meiosis Rye telomeric knobs Telomere cluster when condensation has not begun Telomere cluster (pairing) when condensation has begun

What is the Ph 1 ? Cloning the issues • No natural variation in Ph 1 phenotype- Can’t create a segregating populations, the starting point of all previous positional cloning projects- Can’t score thousands of plants for this phenotype , 5000 plants would be 150, 000 metaphase spreads • Variation only occurs with dosage • EMS treatments don’t yield mutants • But X-Ray and fast neutron irradiation do-A single deletion (ph 1 b) of the locus 70 Mb in size- • “Ph 1 locus” arose on polyploidisation not present in diploids- it is specific to 5 B • Heterochromatin compensates for Ph 1 • Ph 1 could be a multigene family or heterochromatin or both • But wheat is 5 times larger than the human genome

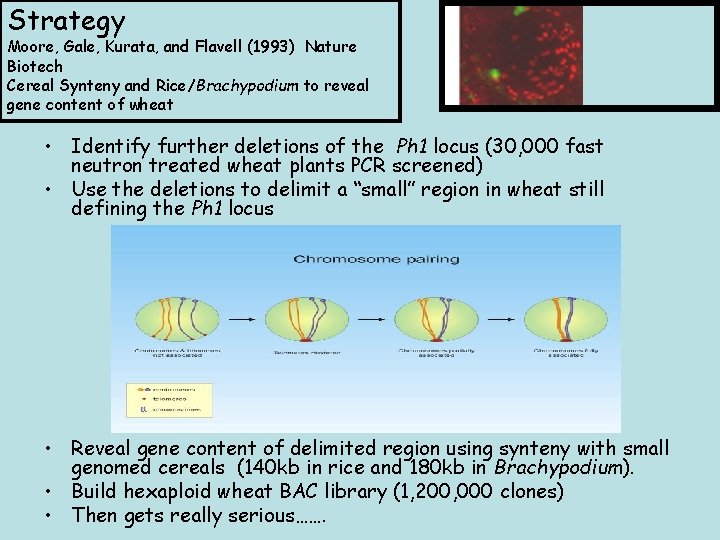
Strategy Moore, Gale, Kurata, and Flavell (1993) Nature Biotech Cereal Synteny and Rice/Brachypodium to reveal gene content of wheat • Identify further deletions of the Ph 1 locus (30, 000 fast neutron treated wheat plants PCR screened) • Use the deletions to delimit a “small” region in wheat still defining the Ph 1 locus • Reveal gene content of delimited region using synteny with small genomed cereals (140 kb in rice and 180 kb in Brachypodium). • Build hexaploid wheat BAC library (1, 200, 000 clones) • Then gets really serious…….

2. 5 Mb Contig Brachypodium/ Rice 5 B Ph 1 regions region

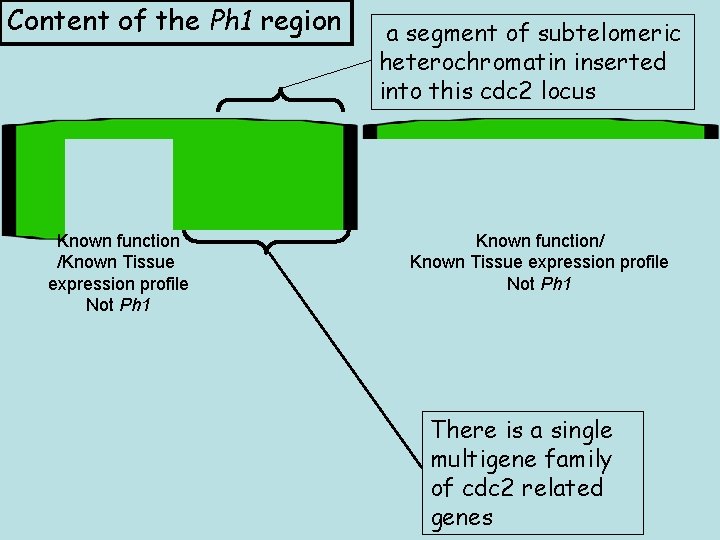
Content of the Ph 1 region Known function /Known Tissue expression profile Not Ph 1 a segment of subtelomeric heterochromatin inserted into this cdc 2 locus Known function/ Known Tissue expression profile Not Ph 1 There is a single multigene family of cdc 2 related genes

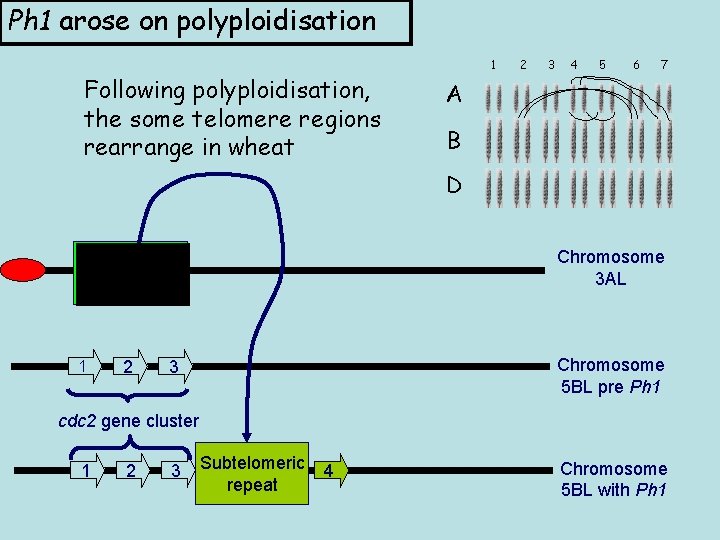
Ph 1 arose on polyploidisation Following polyploidisation, the some telomere regions rearrange in wheat 1 2 3 4 5 6 7 A B D Subtelomeric repeat Chromosome 3 AL 2 Chromosome 5 BL pre Ph 1 1 3 cdc 2 gene cluster 1 2 3 Subtelomeric repeat 4 Chromosome 5 BL with Ph 1

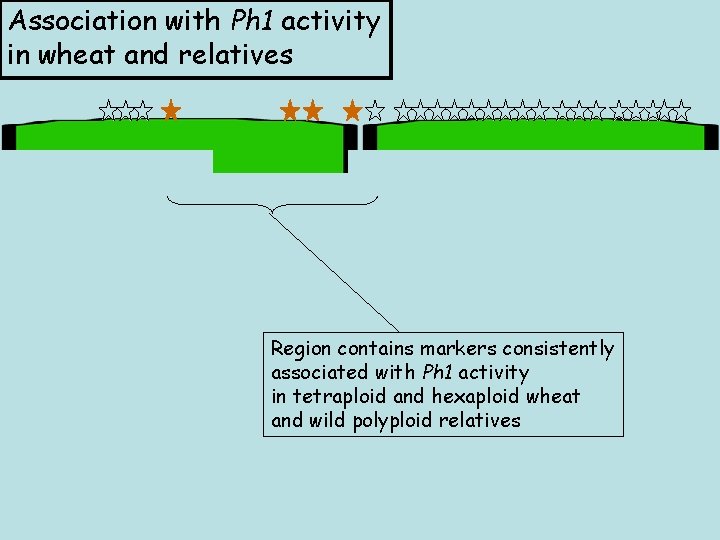
Association with Ph 1 activity in wheat and relatives Region contains markers consistently associated with Ph 1 activity in tetraploid and hexaploid wheat and wild polyploid relatives

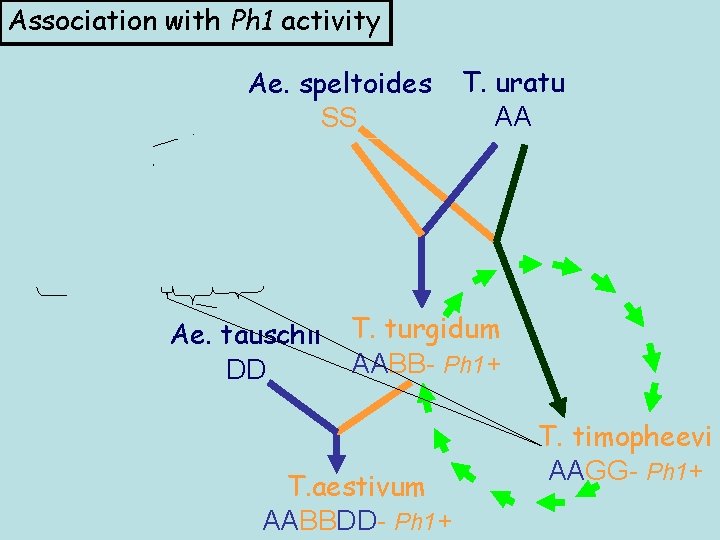
Association with Ph 1 activity Ae. speltoides SS T. uratu AA Ae. tauschii T. turgidum AABB- Ph 1+ DD T. aestivum AABBDD- Ph 1+ T. timopheevi AAGG- Ph 1+

Ph 1 story Summary • We know the cell biological basis of Ph 1 activity • We know the structure of the Ph 1 “locus” • We know that there is 5 B specific expression from within the locus • But we don’t yet know precisely how the locus is working • We need to be able to dissect the components of the locus using small deletions (which may or may be not technically feasible using X-Ray treatment)

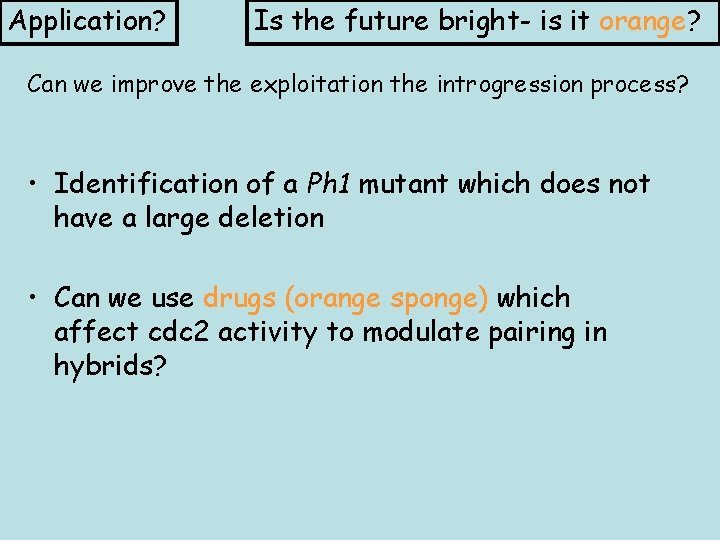
Application? Is the future bright- is it orange? Can we improve the exploitation the introgression process? • Identification of a Ph 1 mutant which does not have a large deletion • Can we use drugs (orange sponge) which affect cdc 2 activity to modulate pairing in hybrids?

Acknowledgements Collaborators Mike Gale -Markers Peter Shaw-Cell biology RGP-Japan - Rice INRA- Evry-BACs Dupont-Sequencing Syngenta-Rush work David Baulcombe si. RNA Cell Biology Luis Aragon Alcaide Enrique (Fadri) Martinez-Perez Pilar Prieto Aranda Mike Wanous Thomas Haizel Physical mapping etc Tracie Foote Michael Roberts Species used Terry Miller Wheat (meiosis and genomics) Steve Reader Rice (meiosis and sequence data) Brachypodium (centromeres and Simon Griffiths sequence data) Sebastien Allouis Luzula , a rush (meiosis and centromeres) Rebecca Sharp Rye (meiosis) Kath Mortimer Isabelle Bertin
 Odplewianie orkiszu
Odplewianie orkiszu Colbert wheat thins
Colbert wheat thins Wheat
Wheat Matthew 13 37
Matthew 13 37 Wheat germ dna factory
Wheat germ dna factory Accelerate wheat
Accelerate wheat Tri a 19
Tri a 19 Flowers in poaceae are
Flowers in poaceae are Quesrcus
Quesrcus Some natural resources such as wheat and cattle are
Some natural resources such as wheat and cattle are What is primary and secondary processing
What is primary and secondary processing Wheat expression browser
Wheat expression browser The wheat and tares
The wheat and tares Wheat
Wheat Wheat adalah
Wheat adalah Food and nutrition 2 state test review
Food and nutrition 2 state test review Antioxidants means
Antioxidants means Grassland wheat
Grassland wheat +micronutrient +fertilizer +wheat
+micronutrient +fertilizer +wheat Australian wheat board
Australian wheat board I could sleep forever figurative language
I could sleep forever figurative language