Eran Yanowski Eran Hornsteins Monitor drug impact on













- Slides: 23

Eran Yanowski, Eran Hornstein’s: Monitor drug impact on the transcriptome of mouse beta cells (primary and cell-line) using Transeq/RNA-Seq 17. 08. 15 Report I

SR 60 Overview of basic parameters of your NGS run (per sample): samples origin: Mouse Beta cells, line

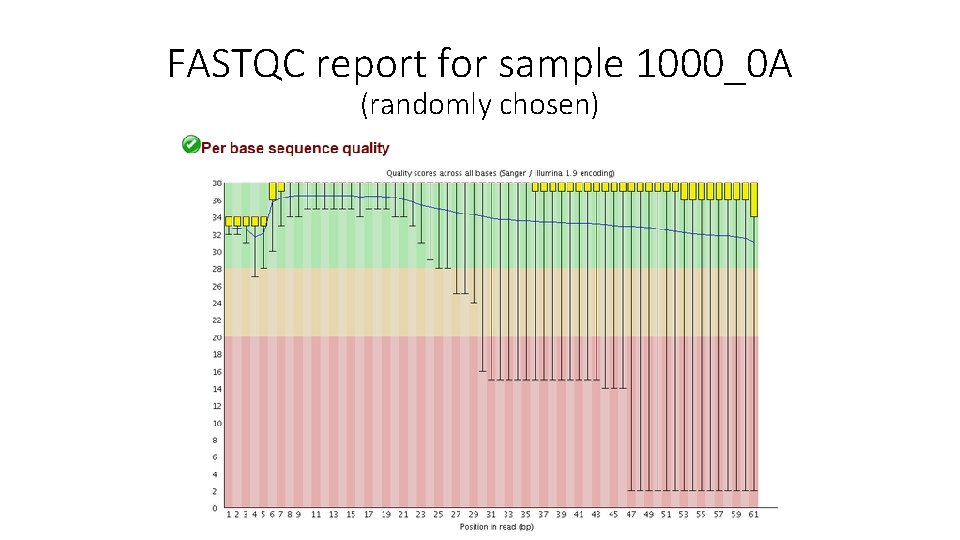
FASTQC report for sample 1000_0 A (randomly chosen)

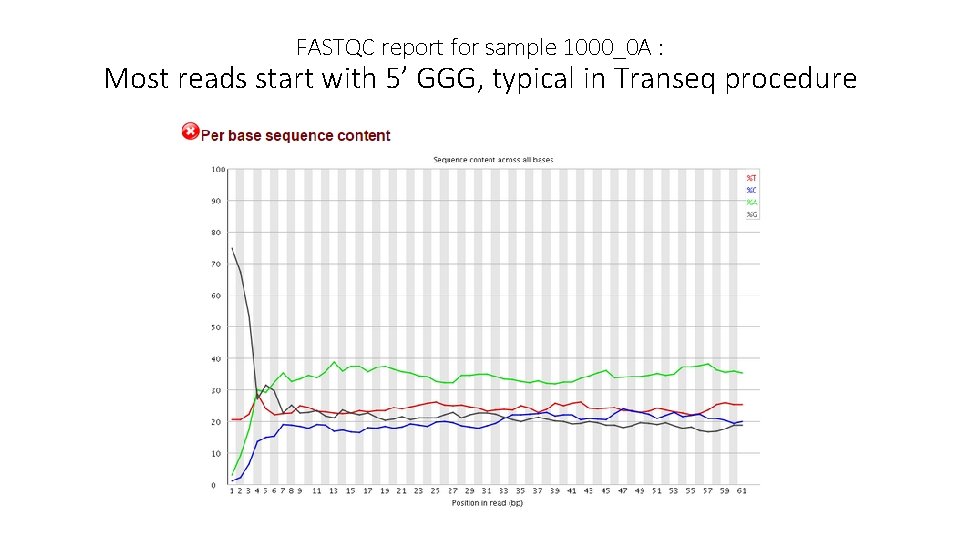
FASTQC report for sample 1000_0 A : Most reads start with 5’ GGG, typical in Transeq procedure

FASTQC report for sample : overrepresentation of Ins 2 reads

Pre-Pipeline data processing Þ Need to remove 5’GGG sequences ÞNeed to remove reads containing either poly-A or poly-T

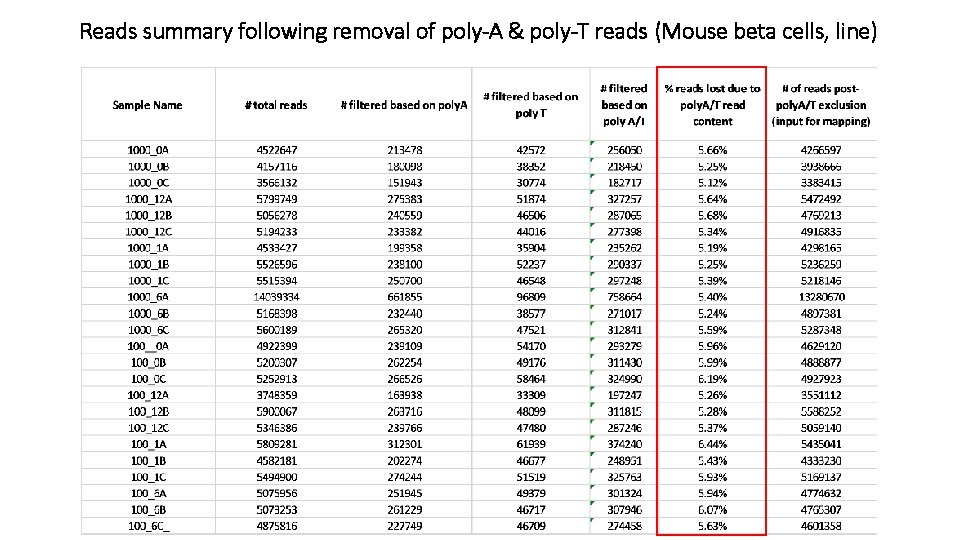
Reads summary following removal of poly-A & poly-T reads (Mouse beta cells, line)

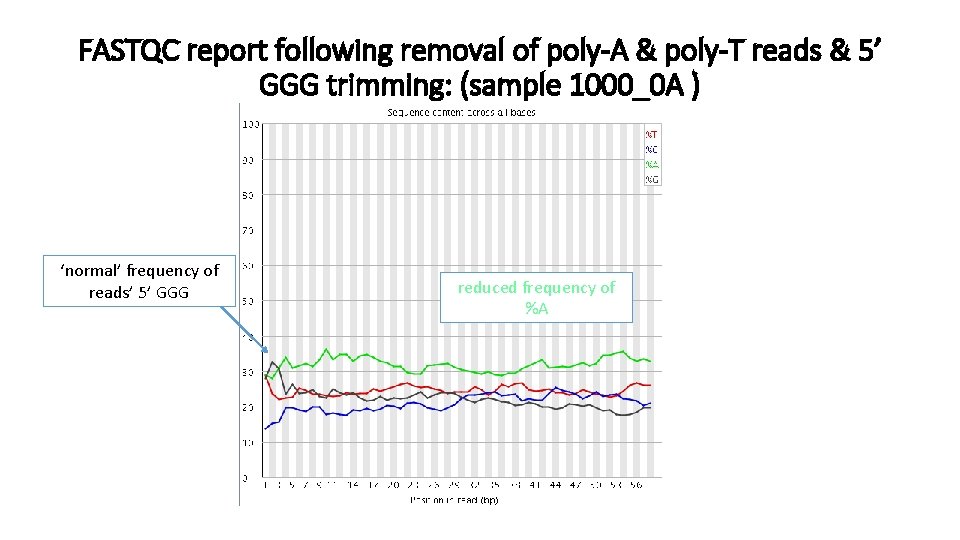
FASTQC report following removal of poly-A & poly-T reads & 5’ GGG trimming: (sample 1000_0 A ) ‘normal’ frequency of reads’ 5’ GGG reduced frequency of %A

Data processing (pipeline) workflow (done using Mouse_mm 9_v 1 base repository) 1. If each sample has more than one fastq file (per sequencing read) then fastq files merging-step is performed 2. Transeq Reads pre-processing (5’ GGG trimming & poly. A & poly. T removal) 3. Processed-reads Mapping (using Top. Hat) 4. TES reads coverage profile (Transeq protocol QC step) 5. Reads Count (per 3’UTR) (using HTSeq-count) 6. Data Normalization and Differential Gene Expression (using DESeq 2) 7. QC: Principal Component Analysis (PCA) & Hierarchical Clustering

Reads mapping summary_Exp 4_1_Mouse Beta cells, line

Reads mapping summary_Exp 4_1_Mouse Beta cells, line: 23 -24% of total reads count were mapped to Ins 2/Ins 1 genes

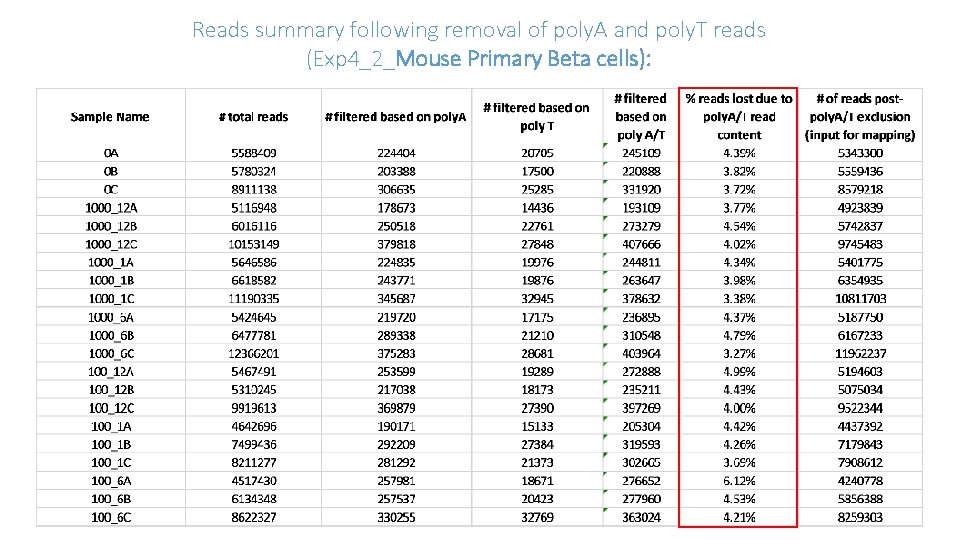
Reads summary following removal of poly. A and poly. T reads (Exp 4_2_Mouse Primary Beta cells):

Reads mapping summary_Exp 4_2_Mouse Primary Beta cells:

Reads mapping summary_Exp 4_2_Mouse Primary Beta cells: % of total reads count were mapped to Ins 2/Ins 1 genes

Hierarchical clustering: Mouse beta cells, line Drug con=100 Drug Con=1000

Hierarchical clustering: Mouse Primary beta cells Drug con=100 Separated by processing day: A/B/C ? Drug Con=1000

• Differential gene expression data is assessed by DESeq 2 • DESeq output is summarized in a single sheet per experiment • Genes differentially-expressed during each time series were called by two independent means: I. Using pairwise comparison vs. time zero II. Using a tool named: ma. Sig. Pro

RNA-Seq drug dose response (1000 and 100)/ time series (0, 1, 6 and 12 hrs) gene filtering criteria • The data filters used during the last analysis performed (12. 08. 15): I. ma. Sig. Pro: • Norm. Counts of genes meeting Max. Raw. Count>50 served as input; • ma. Sig. Pro output was further filtered against potential outlier genes (flagged by ma. Sig. Pro) • Genes showing FC greater than 1. 5 (at least in one of the paired-comparisons); • By default ma. Sig. Pro requires (BH) adjusted p-value <0. 05 II. Pairwise comparison criteria: Max. Raw. Count>50, adjusted-p-value<0. 05 and FC greater than 1. 5 (at least in one of the paired-comparisons);

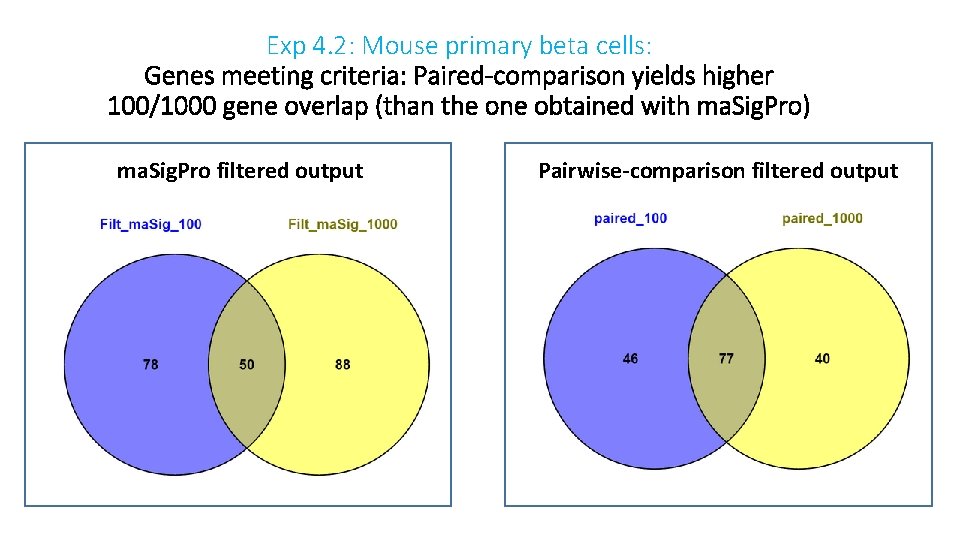
Exp 4. 2: Mouse primary beta cells: Genes meeting criteria: Paired-comparison yields higher 100/1000 gene overlap (than the one obtained with ma. Sig. Pro) ma. Sig. Pro filtered output Pairwise-comparison filtered output

Exp 4. 2: Mouse primary beta cells: Most of ma. Sig. Pro shared-genes are included in the group of paired shared genes To determine which output is preferred data validation using an orthogonal method is essential

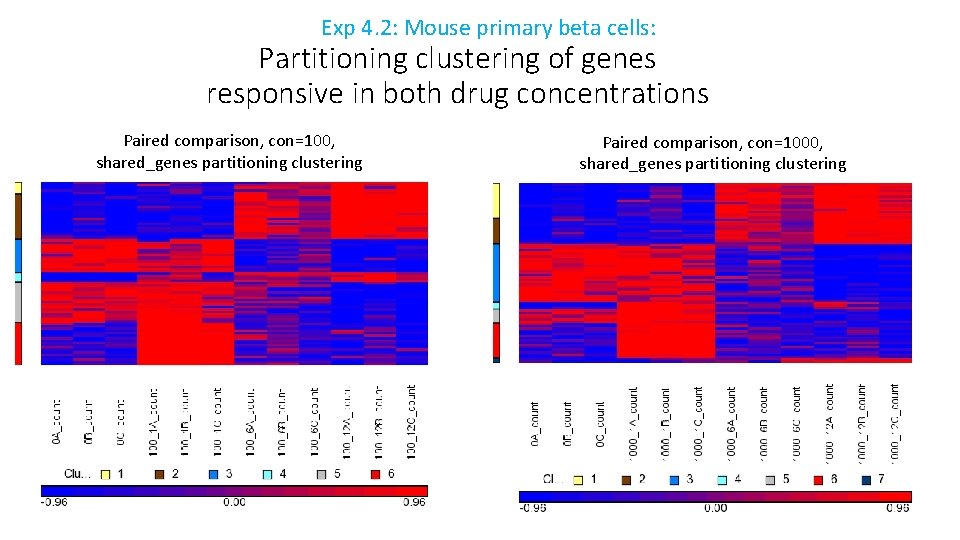
Exp 4. 2: Mouse primary beta cells: Partitioning clustering of genes responsive in both drug concentrations Paired comparison, con=100, shared_genes partitioning clustering Paired comparison, con=1000, shared_genes partitioning clustering

Exp 4. 1: Mouse Beta Cells, cell line • Generally this experiment yielded less significant results • When applying the same filters used for the primary beta-cells datasets, very few genes pass; • The possibility of using p-value (instead of adjusted-p-value should be tested by the investigator) • The level of 100/1000 intersection (shared-genes) is lower here compared to the one observed in the primary cells experiment

Venn diagram of unfiltered ma. Sig. Pro outputs of both the primary (100, 1000) and the cell-line Tran. Seq datasets • Low overall intersection between the primary and the cell-line ‘significant’ genes; • Relatively low intersection between the two drug concentrations tested on the beta cell line
 Eran (fueron/eran) las doce.
Eran (fueron/eran) las doce. Methods of adulteration of crude drugs
Methods of adulteration of crude drugs Quienes eran los publicanos
Quienes eran los publicanos Achi brandt
Achi brandt Dr ronald lev
Dr ronald lev Colosenses 2 12-14 explicacion
Colosenses 2 12-14 explicacion Eran segal
Eran segal Civilizacion greco romana
Civilizacion greco romana Yaniv segal
Yaniv segal Intentalo fueron/eran las doce
Intentalo fueron/eran las doce Malquesis y quelosis vestimenta
Malquesis y quelosis vestimenta Quiénes eran los sofistas
Quiénes eran los sofistas Tnica
Tnica Martin luther and his wife
Martin luther and his wife Características de atenas
Características de atenas Picunches
Picunches Eledad
Eledad Que eran los polis
Que eran los polis Que es lo que le agrada a dios
Que es lo que le agrada a dios Quienes eran los sofistas
Quienes eran los sofistas Como eran las viviendas de los comechingones
Como eran las viviendas de los comechingones Eran nuestras dolencias las que llevaba
Eran nuestras dolencias las que llevaba Virtudes de job
Virtudes de job Versiculos de primera comunion
Versiculos de primera comunion