Types of Chromosome Mutations Chromosome Mutations Changes in


- Slides: 13

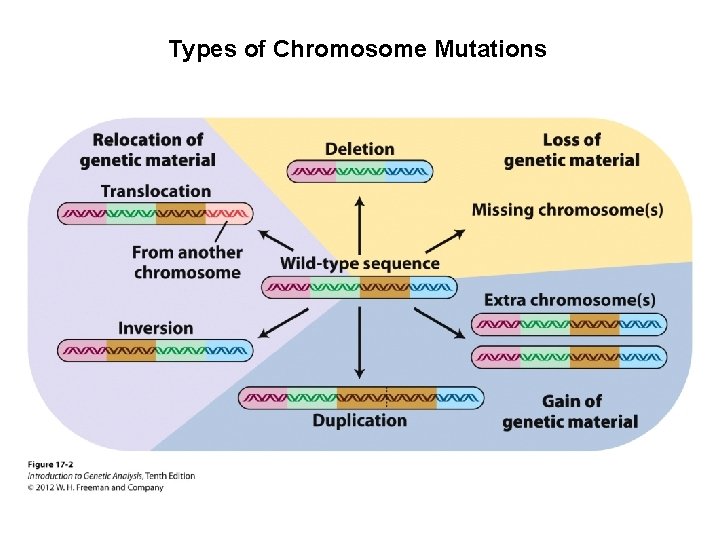
Types of Chromosome Mutations

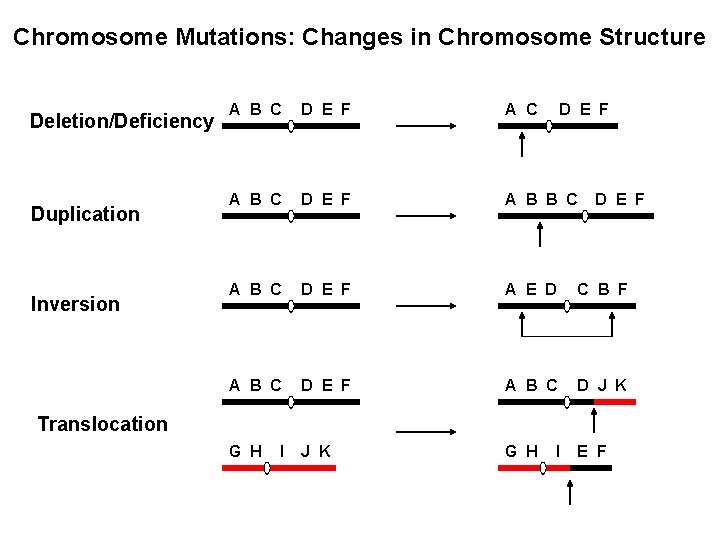
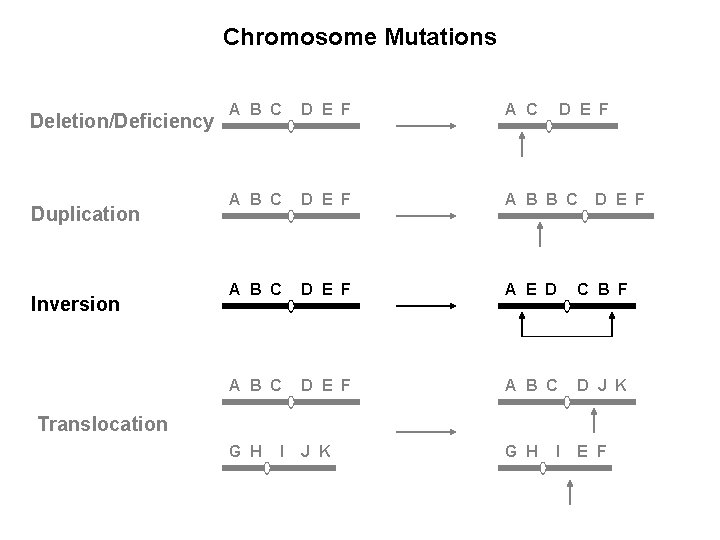
Chromosome Mutations: Changes in Chromosome Structure Deletion/Deficiency Duplication Inversion A B C D E F A B B C A B C D E F A E D C B F A B C D E F A B C D J K G H E F D E F Translocation I I

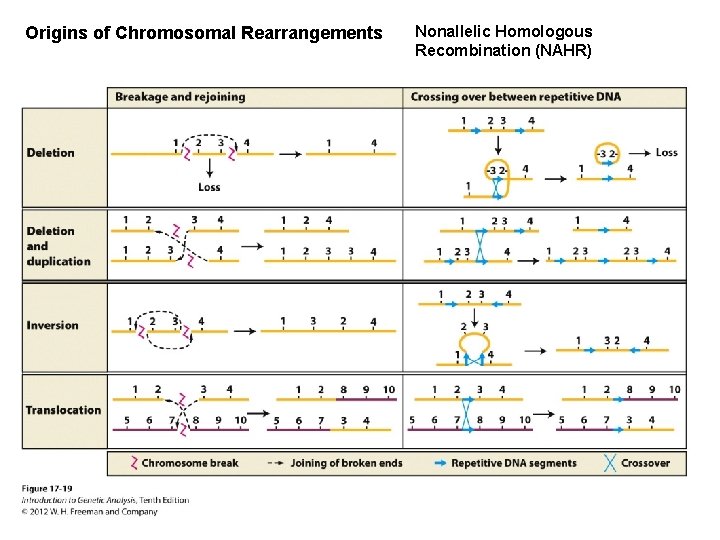
Origins of Chromosomal Rearrangements Nonallelic Homologous Recombination (NAHR)

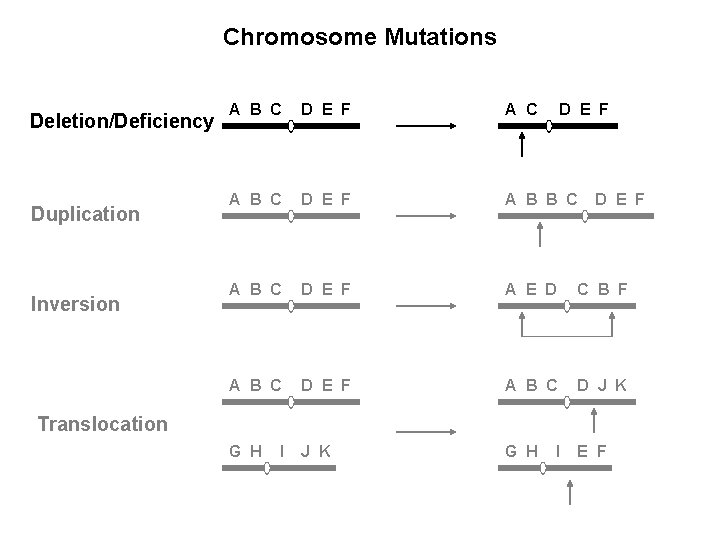
Chromosome Mutations Deletion/Deficiency Duplication Inversion A B C D E F A B B C A B C D E F A E D C B F A B C D E F A B C D J K G H E F D E F Translocation I I

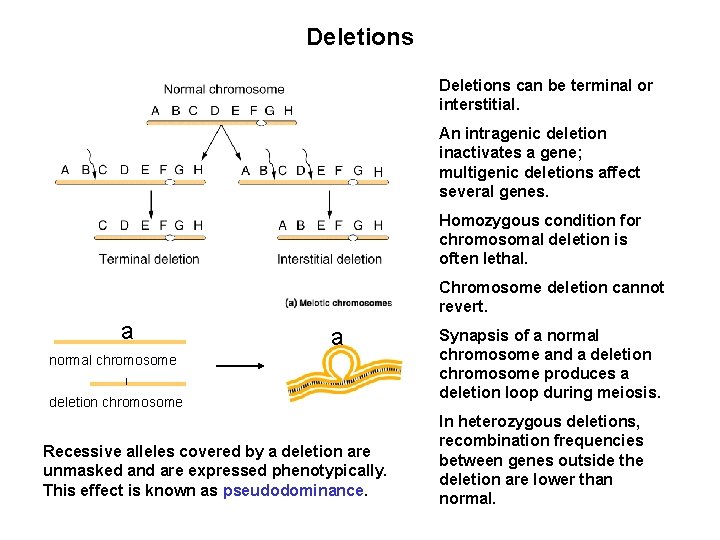
Deletions can be terminal or interstitial. An intragenic deletion inactivates a gene; multigenic deletions affect several genes. Homozygous condition for chromosomal deletion is often lethal. Chromosome deletion cannot revert. a a normal chromosome deletion chromosome Recessive alleles covered by a deletion are unmasked and are expressed phenotypically. This effect is known as pseudodominance. Synapsis of a normal chromosome and a deletion chromosome produces a deletion loop during meiosis. In heterozygous deletions, recombination frequencies between genes outside the deletion are lower than normal.

Deletions Cancer cells often harbor chromosome mutations. Cells of a tumor do not always show the same chromosome mutation. Chromosome-specific deletions are associated with certain tumors.

Chromosome Mutations Deletion/Deficiency Duplication Inversion A B C D E F A B B C A B C D E F A E D C B F A B C D E F A B C D J K G H E F D E F Translocation I I

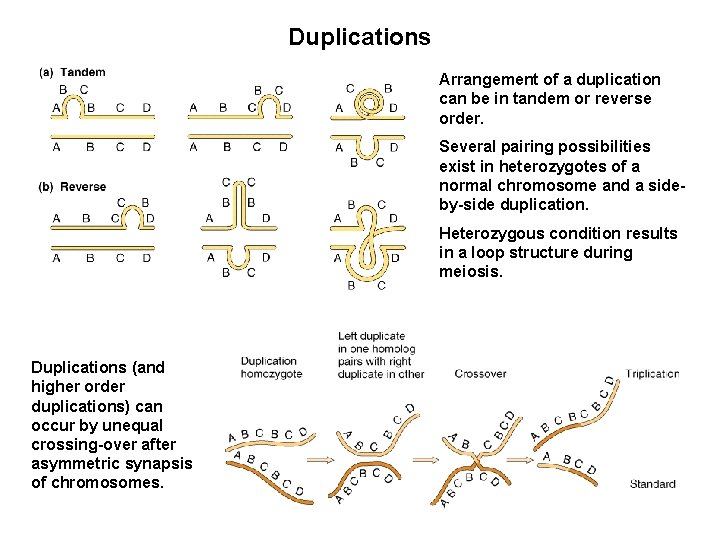
Duplications Arrangement of a duplication can be in tandem or reverse order. Several pairing possibilities exist in heterozygotes of a normal chromosome and a sideby-side duplication. Heterozygous condition results in a loop structure during meiosis. Duplications (and higher order duplications) can occur by unequal crossing-over after asymmetric synapsis of chromosomes.

Chromosome Mutations Deletion/Deficiency Duplication Inversion A B C D E F A B B C A B C D E F A E D C B F A B C D E F A B C D J K G H E F D E F Translocation I I

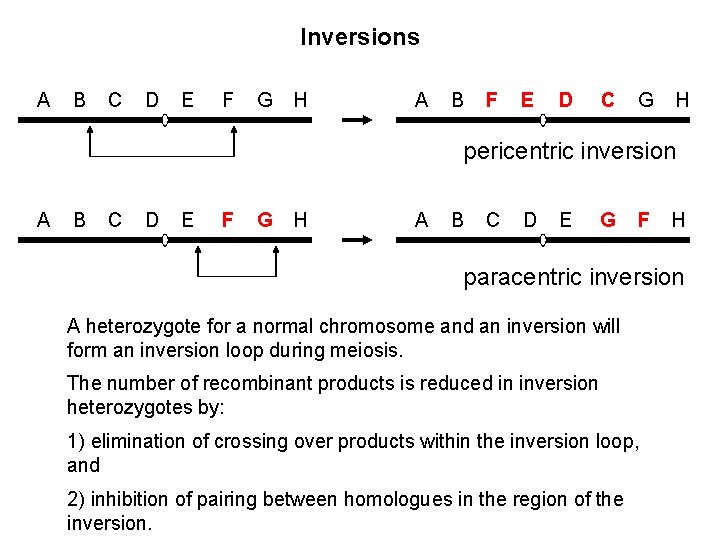
Inversions A B C D E F G H A B F E D C G H pericentric inversion A B C D E F G H A B C D E G F H paracentric inversion A heterozygote for a normal chromosome and an inversion will form an inversion loop during meiosis. The number of recombinant products is reduced in inversion heterozygotes by: 1) elimination of crossing over products within the inversion loop, and 2) inhibition of pairing between homologues in the region of the inversion.

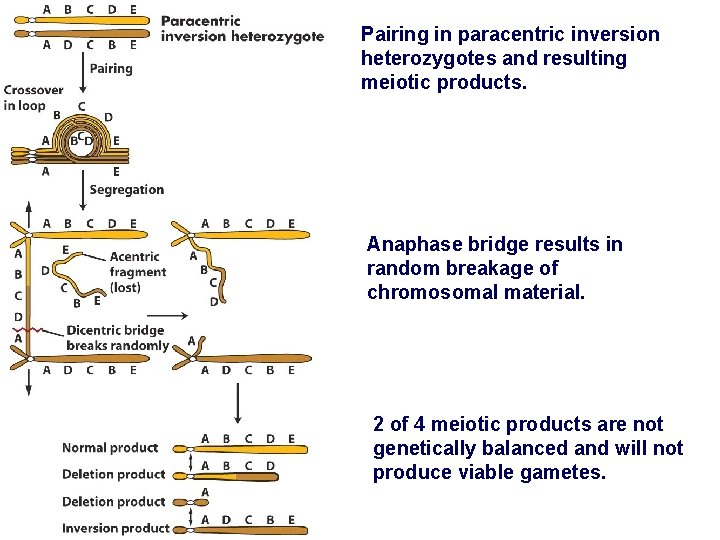
Pairing in paracentric inversion heterozygotes and resulting meiotic products. Anaphase bridge results in random breakage of chromosomal material. 2 of 4 meiotic products are not genetically balanced and will not produce viable gametes.

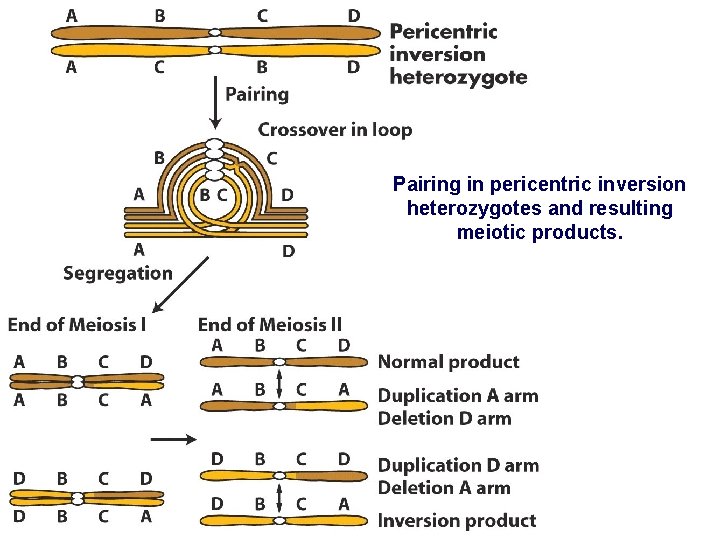
Pairing in pericentric inversion heterozygotes and resulting meiotic products.

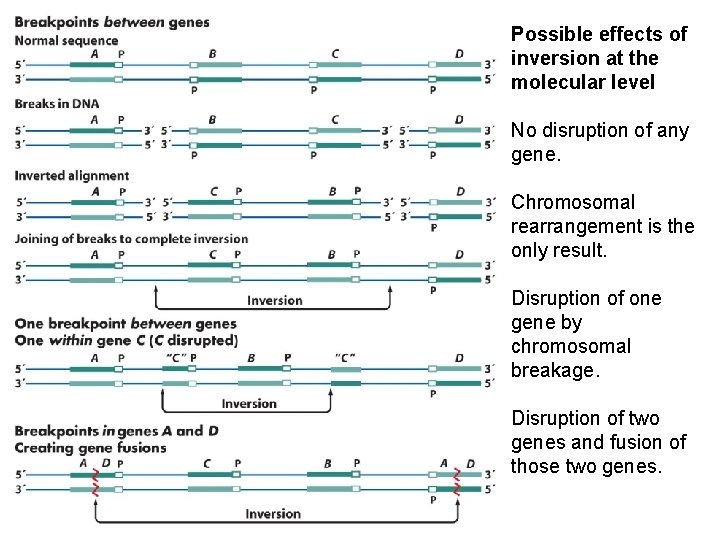
Possible effects of inversion at the molecular level No disruption of any gene. Chromosomal rearrangement is the only result. Disruption of one gene by chromosomal breakage. Disruption of two genes and fusion of those two genes.