Tengcha generic middleware for retrieving data from Chado








- Slides: 11

Tengcha – generic middleware for retrieving data from Chado Justin Reese GMOD Meeting April 5, 2012

Summary Tengcha is a plug-in to the Trellis framework that allows data to be read from Chado Written in Java Tengcha is used by Web. Apollo to read data from our Chado db’s to help people manually annotate Tengcha can be used as generic middleware to: read data from Chado dbs output as Das or Jbrowse style JSON Source code lives here on Google code: https: //genomancer. googlecode. com/svn/trunk

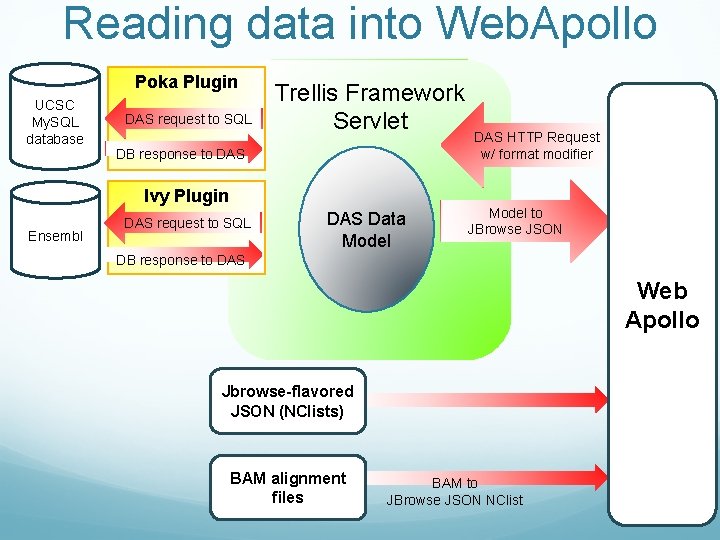
Reading data into Web. Apollo Poka Plugin UCSC My. SQL database DAS request to SQL Trellis Framework Servlet DB response to DAS Ivy Plugin Ensembl DAS request to SQL DAS Data Model DAS HTTP Request w/ format modifier Model to JBrowse JSON DB response to DAS Web Apollo Jbrowse-flavored JSON (NClists) BAM alignment files BAM to JBrowse JSON NClist

Problem – lots of data in Chado databases Much of our (and others’) data lives in Chado databases: protein alignments gene calls RNAseq data/expression data etc. Could convert data to JSON and get JBROWSE to handle the data, but it’d be easier if we pulled it directly from Chado database

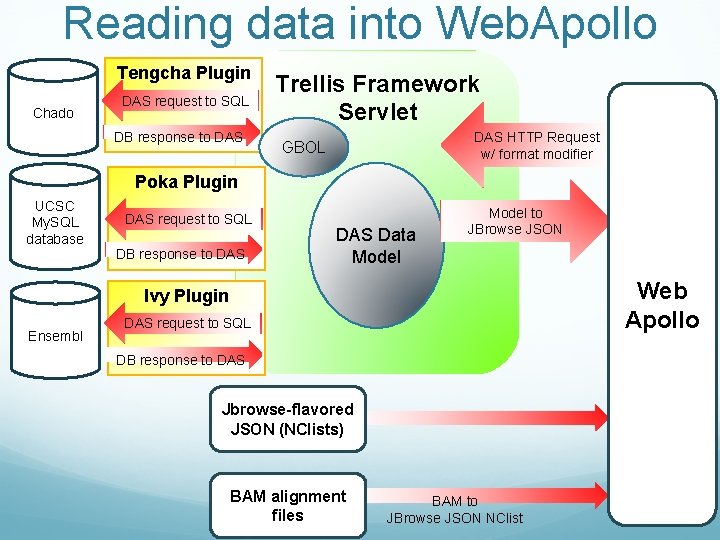
Reading data into Web. Apollo Tengcha Plugin Chado DAS request to SQL DB response to DAS Trellis Framework Servlet DAS HTTP Request w/ format modifier GBOL Poka Plugin UCSC My. SQL database DAS request to SQL DB response to DAS Data Model to JBrowse JSON Web Apollo Ivy Plugin Ensembl DAS request to SQL DB response to DAS Jbrowse-flavored JSON (NClists) BAM alignment files BAM to JBrowse JSON NClist

Tengcha Trellis is a java-based plug-in to Trellis framework Trellis can read data from many places: UCSC (via Poka plug-in) DAS servers (via Ivy plug-in) previously no plug-in to read data from Chado Trellis can output data in a few formats: Das 2 JSON (Jbrowse-flavored JSON) Possibly Das 1 in the future? Design goals: should read data from all standard Chado databases (not just our Chado databases) with data loaded using GMOD bulk loader, with very minimal configuration should be easily configurable to read data from non-standard Chado database should be reasonably fast (Chado is normalized, can be slow…) should be thoroughly unit-tested

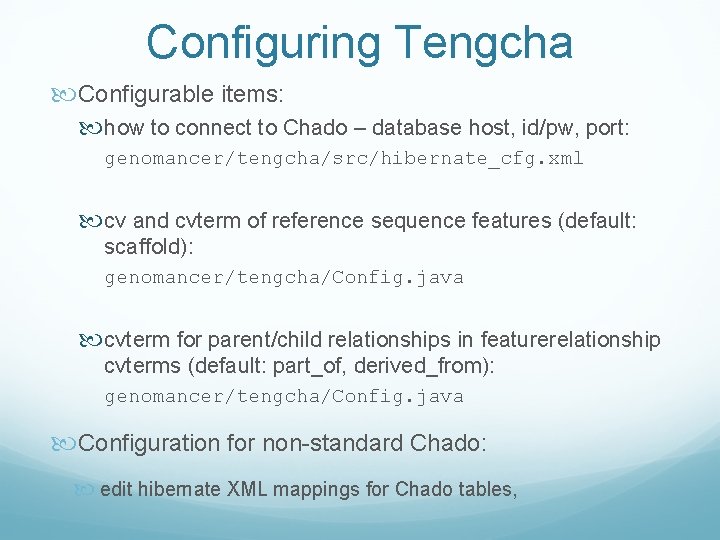
Configuring Tengcha Configurable items: how to connect to Chado – database host, id/pw, port: genomancer/tengcha/src/hibernate_cfg. xml cv and cvterm of reference sequence features (default: scaffold): genomancer/tengcha/Config. java cvterm for parent/child relationships in featurerelationship cvterms (default: part_of, derived_from): genomancer/tengcha/Config. java Configuration for non-standard Chado: edit hibernate XML mappings for Chado tables,

Tengcha as a generic tool for reading from Chado Easy interoperability b/t Chado and anything that speaks Das Output Chado features in Das (XML) Nested-containment lists (JSON) Caching of painful reads (highly configurable caching through hibernate) Java-based, if you like that sort of thing

For the Chado mavens Relevant tables: featureloc featurerelationship analysisfeature cv cvterm If you haven’t altered these, your non-standard Chados should work out of the box…

Live demo Source code lives here on Google code: https: //genomancer. googlecode. com/svn/trunk We’d be glad to help you hook it up to your Chado

������ � LBNL � � Ed Lee Gregg Helt Nomi Harris Suzanna Lewis � UC Berkeley � Mitch Skinner � Rob Buels � Ian Holmes � Georgetown University � � � Chris Childers Justin Reese Mónica Muñoz-Torres Jay Sundaram Christine Elsik