POPULATIONGENETIC STRUCTURE OF A RARE PLANT ARISTOLOCHIA CONTORTA






- Slides: 13

POPULATION-GENETIC STRUCTURE OF A RARE PLANT ARISTOLOCHIA CONTORTA BUNGE (ARISTOLOCHIACEAE) Olga V. Nakonechnaya, Olga G. Koren, Alla B. Kholina, Yuri N. Zhuravlev Institute of Biology and Soil Science, FEB RAS, Vladivostok

rsk mo Pri A. contorta Bunge is a unique herbaceous liana of the Russian Far East, where this plant is represented by a few small fragmented and badly exhausted populations in the southwest part of the Primorsky Krai and in the Jewish Autonomous Region only. In Russia, the species is listed in the Red Book of the Primorsky Krai as a vulnerable species. This plant is uncommon in nature through its sensitivity to destructive changes of habitats. y. K rai Aristolochia contorta Bunge

In addition, A. contorta is a valuable medicine plant and its collection for a herbal remedy contributes to reduction of its natural populations. Now these populations need in protection and restoring. It would be specially important for the saving not only this species, but also for the closely connected with A. contorta relict butterfly Sericinus montela Gray, for which this plant is the single feed source. Sericinus montela Gray

In this work we try to estimate genetic variation and differentiation of the Russian populations of A. contorta using allozyme markers. The following tasks were formulated: 1. To estimate the main parameters of genetic variation for four natural populations of A. contorta from Petrovka, Sukhodol and Razdol’naya rivers basins in Primorsky krai. 2. To estimate variation and differentiation degree among the populations.

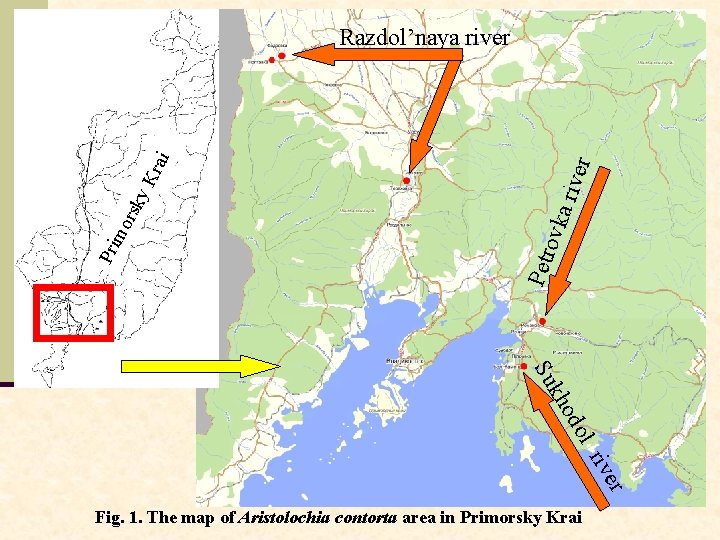
er a riv ovk Petr Pri mo rsk y. K rai Razdol’naya river l do o kh Su er riv Fig. 1. The map of Aristolochia contorta area in Primorsky Krai

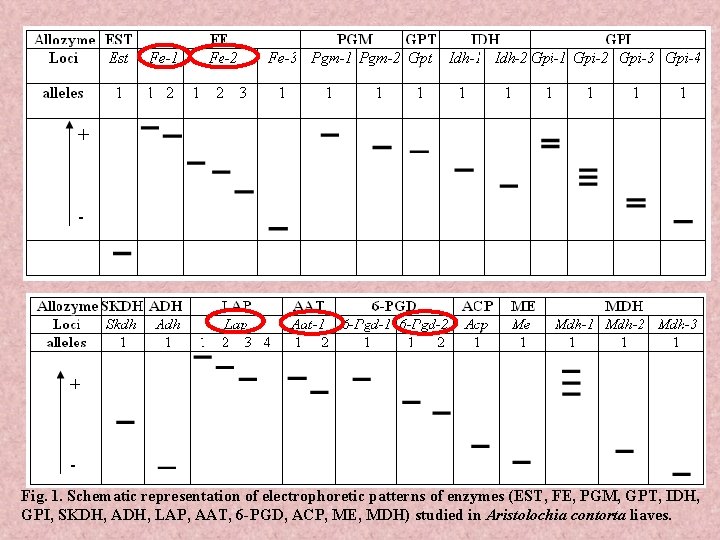
Fig. 1. Schematic representation of electrophoretic patterns of enzymes (EST, FE, PGM, GPT, IDH, GPI, SKDH, ADH, LAP, AAT, 6 -PGD, ACP, ME, MDH) studied in Aristolochia contorta liaves.

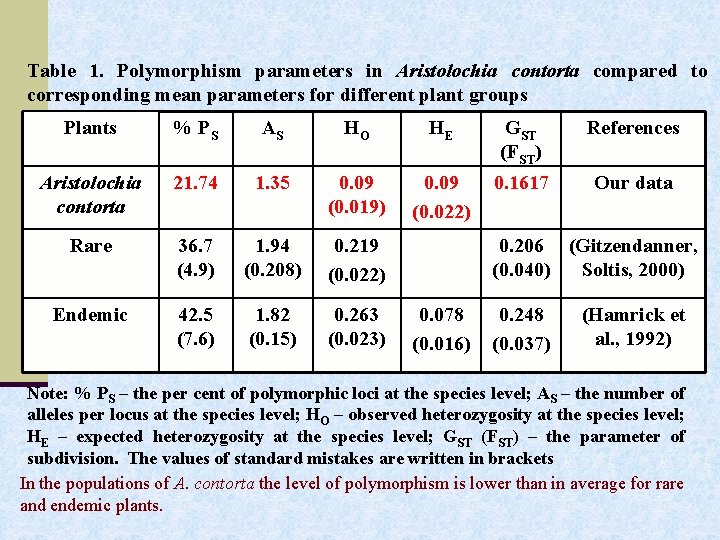
Table 1. Polymorphism parameters in Aristolochia contorta compared to corresponding mean parameters for different plant groups Plants % PS AS НO НE GST (FST) References Aristolochia contorta 21. 74 1. 35 0. 09 (0. 019) 0. 09 (0. 022) 0. 1617 Our data Rare 36. 7 (4. 9) 1. 94 (0. 208) 0. 219 (0. 022) Endemic 42. 5 (7. 6) 1. 82 (0. 15) 0. 263 (0. 023) 0. 206 (Gitzendanner, (0. 040) Soltis, 2000) 0. 078 (0. 016) 0. 248 (0. 037) (Hamrick et al. , 1992) Note: % PS – the per cent of polymorphic loci at the species level; AS – the number of alleles per locus at the species level; НO – observed heterozygosity at the species level; НE – expected heterozygosity at the species level; GST (FST) – the parameter of subdivision. The values of standard mistakes are written in brackets In the populations of A. contorta the level of polymorphism is lower than in average for rare and endemic plants.

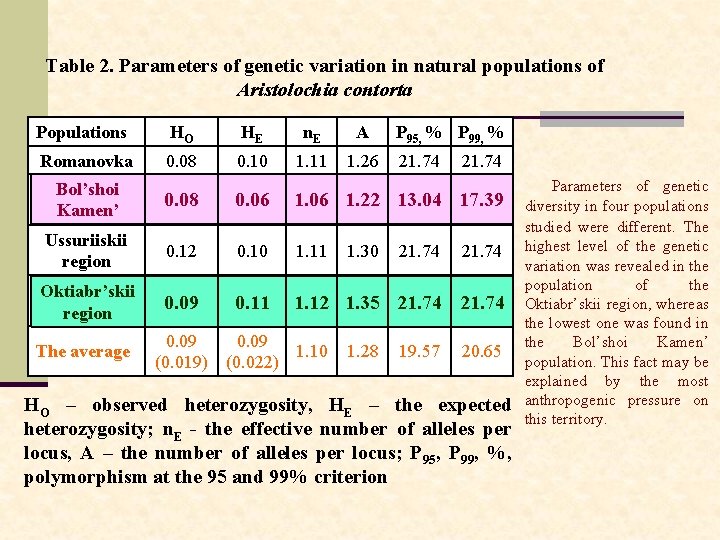
Table 2. Parameters of genetic variation in natural populations of Aristolochia contorta Populations HО HЕ n. Е A Romanovka 0. 08 0. 10 1. 11 1. 26 P 95, % P 99, % 21. 74 Bol’shoi Kamen’ 0. 08 0. 06 1. 22 13. 04 17. 39 Ussuriiskii region 0. 12 0. 10 1. 11 Oktiabr’skii region 0. 09 0. 11 1. 12 1. 35 21. 74 1. 12 The average 0. 09 (0. 019) 0. 09 1. 10 (0. 022) 1. 30 1. 28 21. 74 19. 57 21. 74 20. 65 HO – observed heterozygosity, HE – the expected heterozygosity; n. Е - the effective number of alleles per locus, A – the number of alleles per locus; P 95, P 99, %, polymorphism at the 95 and 99% criterion Parameters of genetic diversity in four populations studied were different. The highest level of the genetic variation was revealed in the population of the Oktiabr’skii region, whereas the lowest one was found in the Bol’shoi Kamen’ population. This fact may be explained by the most anthropogenic pressure on this territory.

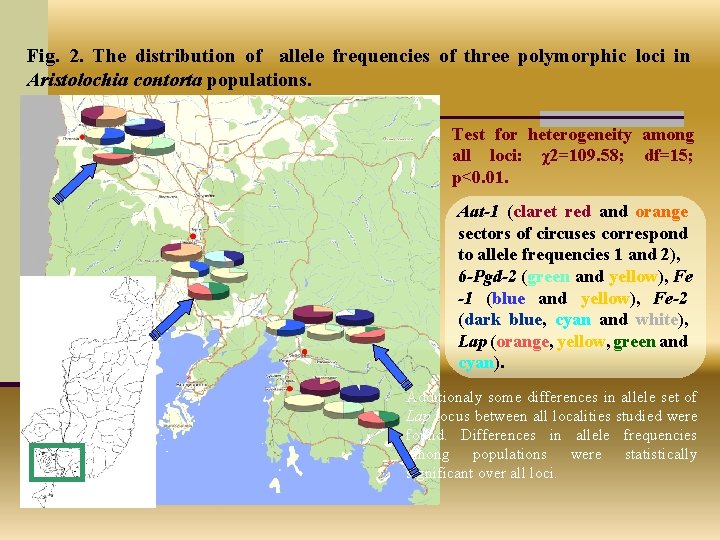
Fig. 2. The distribution of allele frequencies of three polymorphic loci in Aristolochia contorta populations. Test for heterogeneity among all loci: χ2=109. 58; df=15; р<0. 01. Aat-1 (claret red and orange sectors of circuses correspond to allele frequencies 1 and 2), 6 -Pgd-2 (green and yellow), Fe -1 (blue and yellow), Fe-2 (dark blue, cyan and white), Lap (orange, yellow, green and cyan). Additionaly some differences in allele set of Lap locus between all localities studied were found. Differences in allele frequencies among populations were statistically significant over all loci.

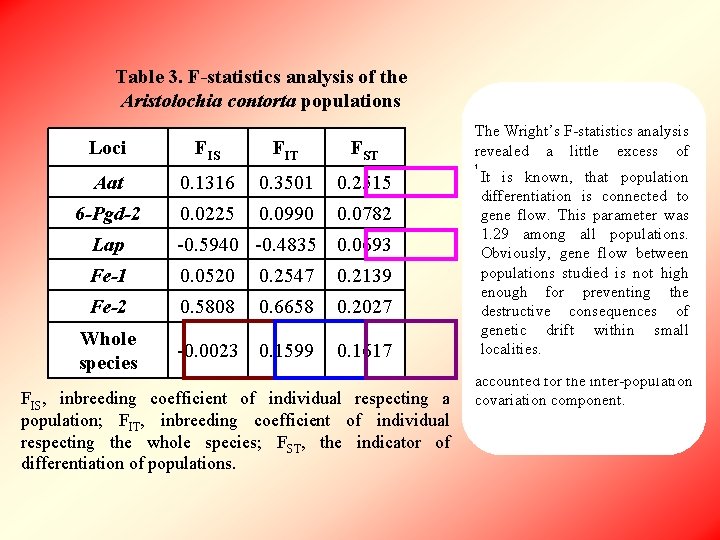
Table 3. F-statistics analysis of the Aristolochia contorta populations Loci FIS FIT FST Aat 0. 1316 0. 3501 0. 2515 6 -Pgd-2 0. 0225 0. 0990 0. 0782 Lap -0. 5940 -0. 4835 0. 0693 Fe-1 0. 0520 0. 2547 0. 2139 Fe-2 0. 5808 0. 6658 0. 2027 -0. 0023 0. 1599 0. 1617 Whole species FIS, inbreeding coefficient of individual respecting a population; FIT, inbreeding coefficient of individual respecting the whole species; FST, the indicator of differentiation of populations. Nem=1. 29 The Wright’s F-statistics analysis revealed a little excess of heterozygotes the populations It is known, in that population and, simultaneously, about differentiation is connected 16% to heterozygote deficiency for the gene flow. This parameter was whole maypopulations. be evidence 1. 29 species. among Itall of. Obviously, subdivisiongene of the species into flow between separate populationssmall studiedreproductive is not high groups. The differentiation enough for preventing index the Fdestructive varied from 0. 0693 to 0. 2515 ST consequences of and was 0. 16 all genetic drift averaged within for small loci. This value says that about localities. 16. 2% of the total variation is accounted for the inter-population covariation component.

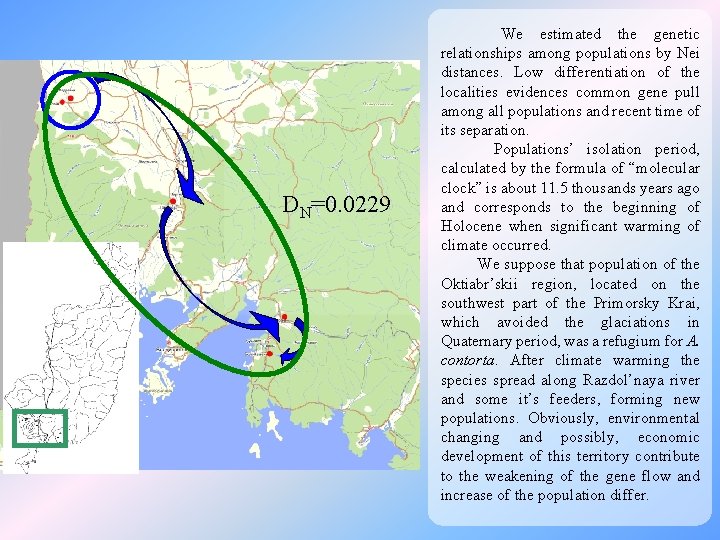
DN=0. 0229 We estimated the genetic relationships among populations by Nei distances. Low differentiation of the localities evidences common gene pull among all populations and recent time of its separation. Populations’ isolation period, calculated by the formula of “molecular clock” is about 11. 5 thousands years ago and corresponds to the beginning of Holocene when significant warming of climate occurred. We suppose that population of the Oktiabr’skii region, located on the southwest part of the Primorsky Krai, which avoided the glaciations in Quaternary period, was a refugium for A. contorta. After climate warming the species spread along Razdol’naya river and some it’s feeders, forming new populations. Obviously, environmental changing and possibly, economic development of this territory contribute to the weakening of the gene flow and increase of the population differ.

The conclusion Thus, this study showed a low level of genetic diversity and high level of genetic differentiation between Russian A. contorta populations. The data supposed that all the populations of A. contorta in Primorsky Krai have common gene pool. Today genetic drift is contributing the main input to the population genetic structure of the A. contorta. The enhancing of genetic drift is connected with reduction of reproductive and effective population sizes under increasing of anthropogenic pressure.

 Calceolaria uniflora
Calceolaria uniflora Duitse pijp giftig
Duitse pijp giftig Phellem cork
Phellem cork Aristolochia salvadorensis
Aristolochia salvadorensis Objective of plant breeding
Objective of plant breeding Plant breeding for disease resistance
Plant breeding for disease resistance Plant introduction in plant breeding
Plant introduction in plant breeding Tronsmo plant pathology and plant diseases download
Tronsmo plant pathology and plant diseases download Tronsmo plant pathology and plant diseases download
Tronsmo plant pathology and plant diseases download Tronsmo plant pathology and plant diseases download
Tronsmo plant pathology and plant diseases download Caribbean cleaners national geographic
Caribbean cleaners national geographic Rare celule epiteliale plate ce inseamna
Rare celule epiteliale plate ce inseamna Marge sur coût variable par unité de ressource rare
Marge sur coût variable par unité de ressource rare Croquis l'eau en espagne une ressource rare sous pression
Croquis l'eau en espagne une ressource rare sous pression






















