Motif finding methods and algorithms Given a set












- Slides: 17

Motif finding methods and algorithms Given a set of n promoters of n coregulated genes, find a motif common to the promoters. Both the PWM and the motif sequences are unknown. Common methods: 1. Enumeration: Simplest case: look at the frequency of all n-mers * Finds Global Optimum since can search entire space 2. EM algorithms (MEME): Iteratively hone in on the most likely motif model 3. Gibbs sampling methods (Align. Ace, Bio. Prospector) Iteratively replace (‘sample’) sites to retrain the matrix 1

New MEME tools: http: //meme. ebi. edu. au/meme/intro. html (create your own Nth order markov background model) http: //meme. sdsc. edu/meme/doc/fasta-get-markov. html 2

Motif finding using the EM algorithm MEME (Bailey & Elkan 1995) http: //meme. sdsc. edu/meme/intro. html EM algorithm: Expectation-Maximization In one run, trains the matrix model and identifies examples of the matrix MEME works by iteratively refining matrix and identifying sites: 1. Estimate motif model a. Start with an n-mer seed (random or specified) b. Build a matrix by incorporating some of background frequencies 2. Identify examples of the model a. For every n-mer in the input set, identify its probability given the matrix model 3. Re-estimate the motif model a. Calculate a new matrix, based on the weighted frequencies of all n-mers in the set 4. Iteratively refine the matrix and identify sites until convergence. 3

Problem: find a 6 -mer motif in 4 sequences S 1: GGCTATTGCAGATGACGAGATGAGGCCCAGACC S 2: GGATGACTTATATAAAGGACGATAAGAGATGAC S 3: CTAGCTCGTTGAGATGCGCTCCCCGCTC S 4: GATGACGGAGTATTAAAGACTCGATGAGTTATACGA 1. MEME uses an initial EM heuristic to estimate the best Starting-point matrix: G A T C 0. 26 0. 24 0. 18 0. 26 0. 25 0. 26 0. 24 0. 26 0. 28 0. 24 0. 25 0. 22 0. 25 0. 23 0. 30 0. 25 0. 27 0. 24 0. 25 0. 27 4

2. MEME scores the match of all 6 -mers to current matrix G GCTATTGCATATGACGAGATGAGGCCCAGACC Here, just consider the underlined 6 -mers, Although in reality all 6 -mers are scored GGATGACTTATATAAAGGACCGTGATAAGAGATTAC CTAGCTCGTTGAGATGCGCTCCCCGCTC GATGACGGAGTATTAAAGACTCGATGAGTTATACGA 5

2. MEME scores the match of all 6 -mers to current matrix G GCTATTGCATATGACGAGATGAGGCCCAGACC GGATGACTTATATAAAGGACCGTGATAAGAGATTAC CTAGCTCGTTGAGATGCGCTCCCCGCTC GATGACGGAGTATTAAAGACTCGATGAGTTATACGA 3. Reestimate the matrix based on the weighted contribution of all 6 mers G A T C 0. 29 0. 24 0. 17 0. 24 0. 30 0. 22 0. 26 0. 27 0. 22 0. 28 0. 18 0. 24 0. 23 0. 33 0. 24 0. 28 0. 24 0. 27 0. 23 0. 28 0. 24 The height of the bases above corresponds to how much that 6 -mer counts in calculating the new matrix 6

MEME scores the match of all 6 -mers to current matrix G GCTATTGCATATGACGAGATGAGGCCCAGACC GGATGACTTATATAAAGGACCGTGATAAGAGATTAC CTAGCTCGTTGAGATGCGCTCCCCGCTC GATGACGGAGTATTAAAGACTCGATGAGTTATACGA 7

G GCTATTGCATATGACGAGATGAGGCCCAGACC GGATGACTTATATAAAGGACCGTGATAAGAGATTAC CTAGCTCGTTGAGATGCGCTCCCCGCTC GATGACGGAGTATTAAAGACTCGATGAGTTATACGA Reestimate the matrix based on the weighted contribution of all 6 mers G A T C 0. 40 0. 20 0. 15 0. 42 0. 24 0. 30 0. 24 0. 46 0. 18 0. 15 0. 30 0. 45 0. 16 0. 15 0. 28 0. 15 0. 20 0. 16 0. 15 0. 24 The height of the bases above corresponds to how much that 6 -mer counts in calculating the new matrix 8

MEME scores the match of all 6 -mers to current matrix GGCTATTGCATATGACGAGATGAGGCCCAGACC GGATGACTTATATAAAGGACCGTGATAAGAGATTAC CTAGCTCGTTGAGATGCGCTCCCCGCTC GATGACGGAGTATTAAAGACTCGATGAGTTATACGA Iterations continue until convergence (ie. numbers don’t change much between iterations) 9

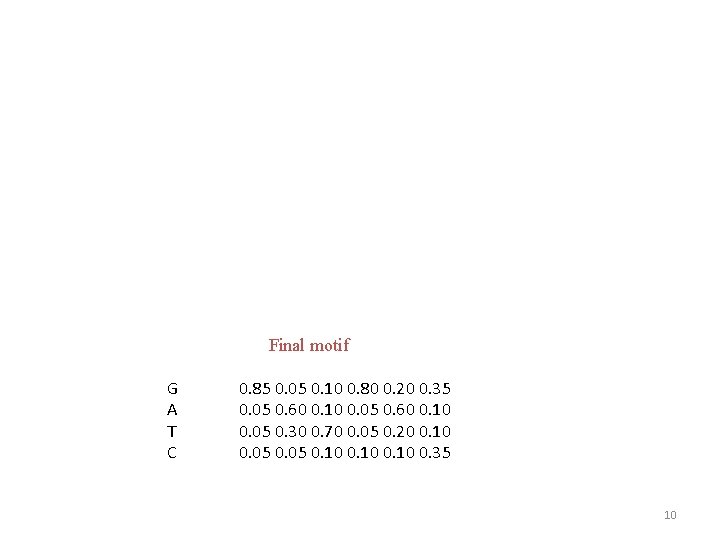
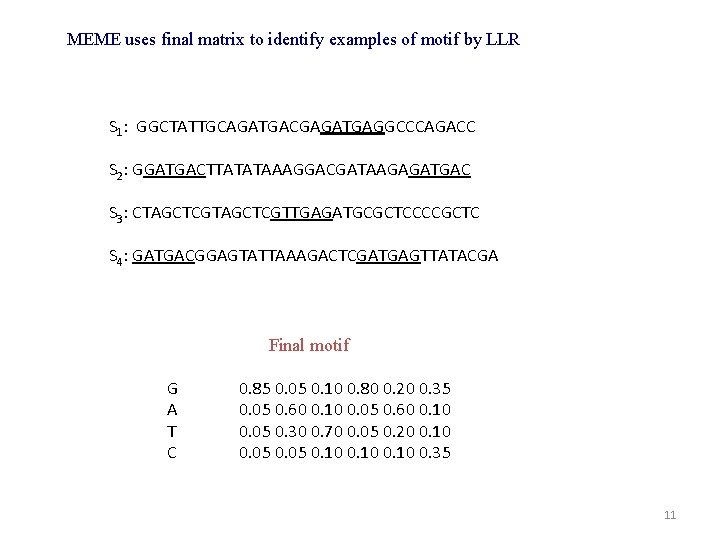
Final motif G A T C 0. 85 0. 05 0. 10 0. 80 0. 20 0. 35 0. 05 0. 60 0. 10 0. 05 0. 30 0. 70 0. 05 0. 20 0. 10 0. 05 0. 10 0. 35 10

MEME uses final matrix to identify examples of motif by LLR S 1: GGCTATTGCAGATGACGAGATGAGGCCCAGACC S 2: GGATGACTTATATAAAGGACGATAAGAGATGAC S 3: CTAGCTCGTTGAGATGCGCTCCCCGCTC S 4: GATGACGGAGTATTAAAGACTCGATGAGTTATACGA Final motif G A T C 0. 85 0. 05 0. 10 0. 80 0. 20 0. 35 0. 05 0. 60 0. 10 0. 05 0. 30 0. 70 0. 05 0. 20 0. 10 0. 05 0. 10 0. 35 11

Motif finding using the EM algorithm MEME (Bailey & Elkan 1995) http: //meme. sdsc. edu/meme/intro. html EM algorithm: Expectation-Maximization In one run, trains the matrix model and identifies examples of the matrix Choice of parameters significantly affects the algorithm -- motif width w -- motif model: - “zoops” = zero-or-one motif per promoter sequence* - “oops” = one-or-more motif per promoter sequence* - “ans” = (“any number of sites”) two-component mixture model (ie. Each w-mer sequence is either an example of the background model or the motif model) -- background model: - simplest case: genomic nucleotide frequencies P(G, A, T, C) - nth-order Markov chain (eg. 2 nd order Markov chain = P(Ai|Ci-1) = P(CA) = dinucleotide frequencies) *These models keep track of which input sequence (promoter) the motif came from, whereas ‘ans’ throws all “w-mers” into a bag 12

Assessing the biological relevance of identified motifs Keep an eye on these features: 1. Bit score (or normalized bit score) Bit score = Information Content at each position 2. Information content profile Real TF binding sites typically show smooth IC profiles 3. Number of input sequences that contain the motif Overfitting: great looking motif but found in only few of the input sequences 4. Nucleotide frequencies Eg. In yeast, AT rich sequences are common … doesn’t necessarily mean they’re not real binding sites 5. Enrichment of motif in the training set compared to genomic bg Our old friend, the hypergeometric distribution. 6. Finding the same consensus with different models or methods 7. Any other nonrandom observation can give you confidence (palindromic motif, nonrandom distribution of motifs in input sequences, etc) 13

Comparing matrices and motifs Tom 1. 2. 3. 4. Pick a scoring function Calculate score for query matrix Q against ALL matrices in database Use those scores to estimate a distribution of scores to turn score into a p-value FDR turns p-value into an E value 14

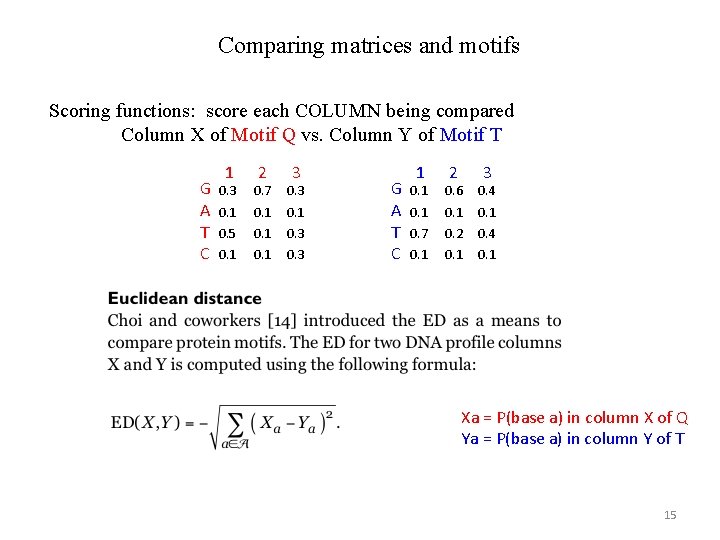
Comparing matrices and motifs Scoring functions: score each COLUMN being compared Column X of Motif Q vs. Column Y of Motif T G A T C 1 2 3 0. 7 0. 3 0. 1 0. 5 0. 1 0. 3 G A T C 1 2 3 0. 1 0. 6 0. 4 0. 1 0. 7 0. 2 0. 4 0. 1 Xa = P(base a) in column X of Q Ya = P(base a) in column Y of T 15

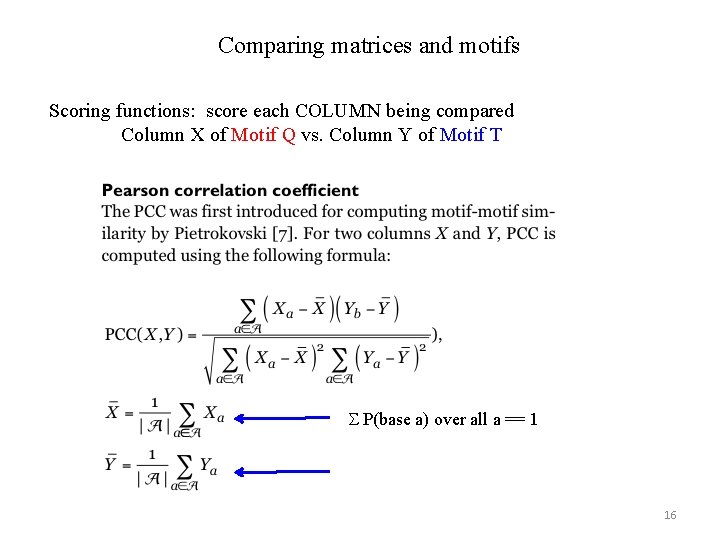
Comparing matrices and motifs Scoring functions: score each COLUMN being compared Column X of Motif Q vs. Column Y of Motif T S P(base a) over all a == 1 16

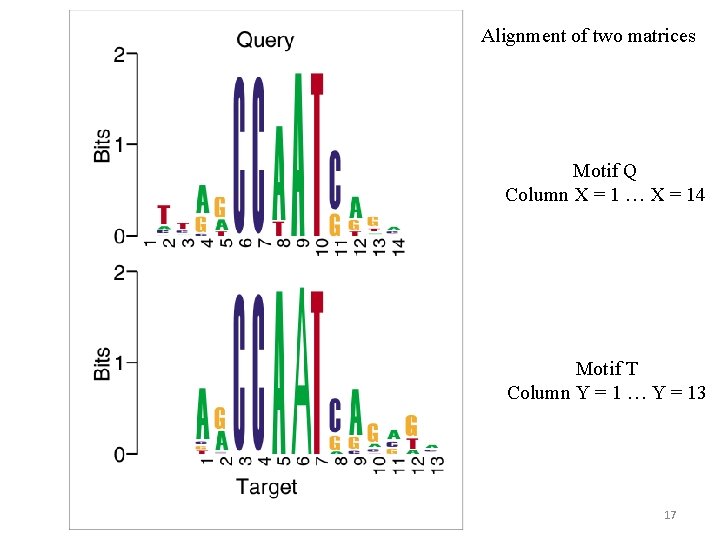
Alignment of two matrices Motif Q Column X = 1 … X = 14 Motif T Column Y = 1 … Y = 13 17