Monday 58 Dr Michael Mc Intosh will introduce






- Slides: 23

Monday 5/8. Dr. Michael Mc. Intosh will introduce psychiatric disease. Bi 1 “Drugs and the Brain” Lecture 18 Thursday, May 4, 2006 A. from m. RNA to Protein B. C. B. Protein degradation 1

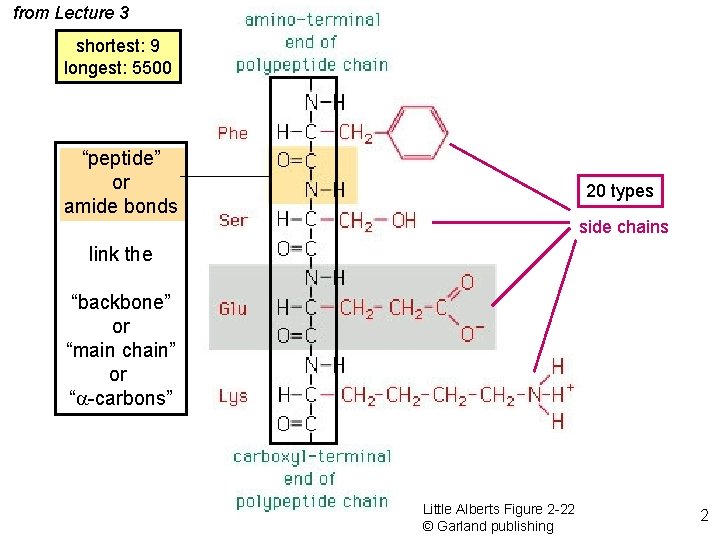
from Lecture 3 shortest: 9 longest: 5500 “peptide” or amide bonds 20 types side chains link the “backbone” or “main chain” or “a-carbons” Little Alberts Figure 2 -22 © Garland publishing 2

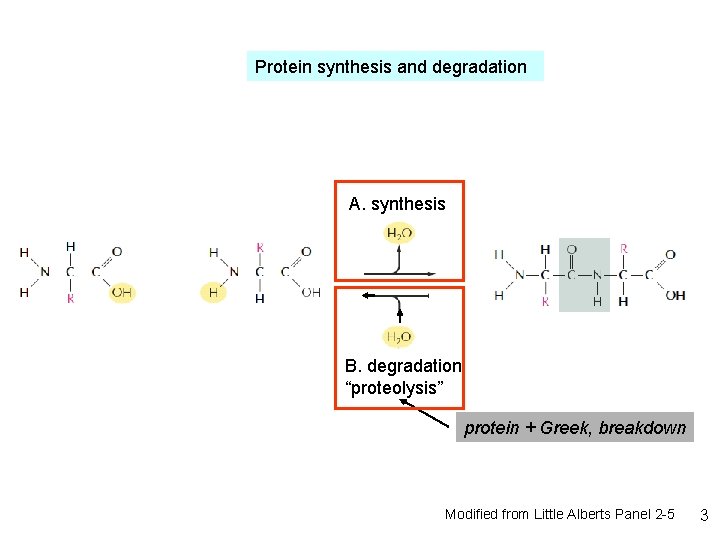
Protein synthesis and degradation A. synthesis B. degradation “proteolysis” protein + Greek, breakdown Modified from Little Alberts Panel 2 -5 3

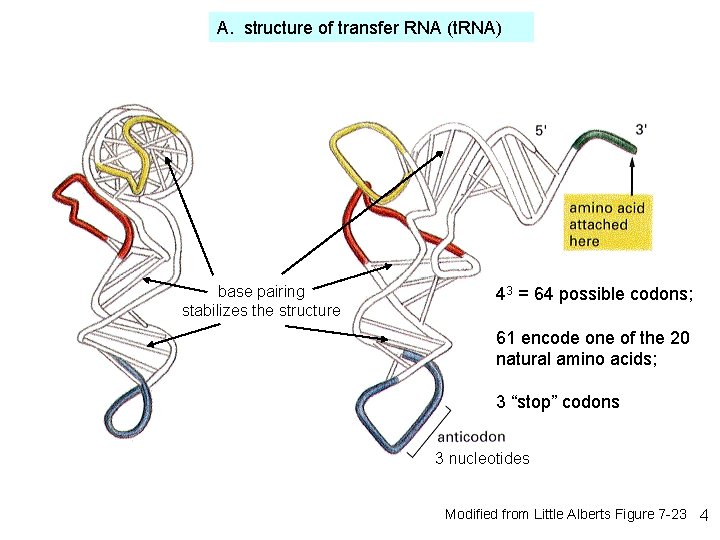
A. structure of transfer RNA (t. RNA) base pairing stabilizes the structure 43 = 64 possible codons; 61 encode one of the 20 natural amino acids; 3 “stop” codons 3 nucleotides Modified from Little Alberts Figure 7 -23 4

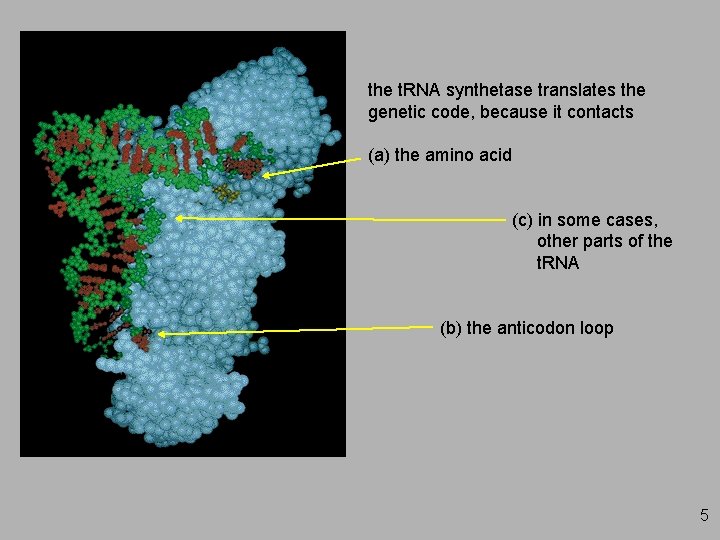
the t. RNA synthetase translates the genetic code, because it contacts (a) the amino acid (c) in some cases, other parts of the t. RNA (b) the anticodon loop 5

Two videos associated with protein synthesis Little Alberts CD-ROM Chapter 7 7. 6, translation 7. 7, polyribosome 6

The discovery of the amber “stop”codon. . . a Caltech story from 1960 Bob Edgar a petri dish Harris Bernstein today © Caltech “My help consisted of flaming a wire loop to sterilize it, then use it to repeatedly pick phage plaques from among hundreds on one petri dish and then inoculate two other petri dishes which had been seeded with host bacteria. . they were pessimistic about the outcome, and we agreed as sort of a joke that if the experiment did work out they would name mutants after my mother. ” 7

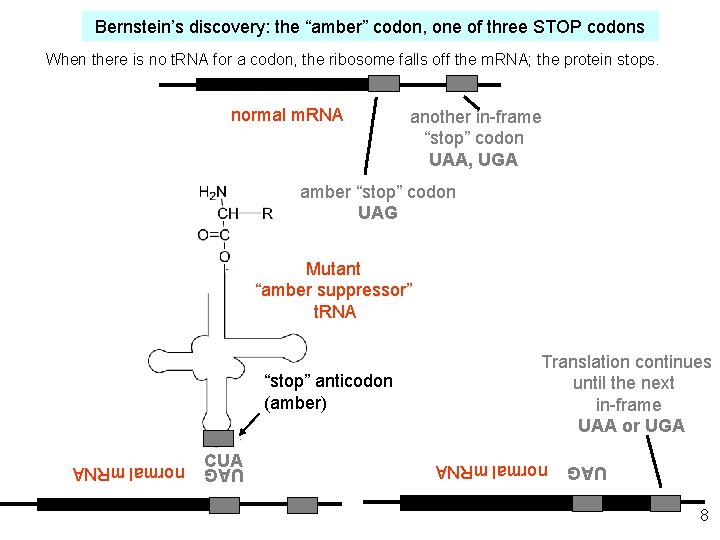
Bernstein’s discovery: the “amber” codon, one of three STOP codons When there is no t. RNA for a codon, the ribosome falls off the m. RNA; the protein stops. normal m. RNA another in-frame “stop” codon UAA, UGA amber “stop” codon UAG Mutant “amber suppressor” t. RNA normal m. RNA CUA UAG “stop” anticodon (amber) Translation continues until the next in-frame UAA or UGA 8 UAG normal m. RNA

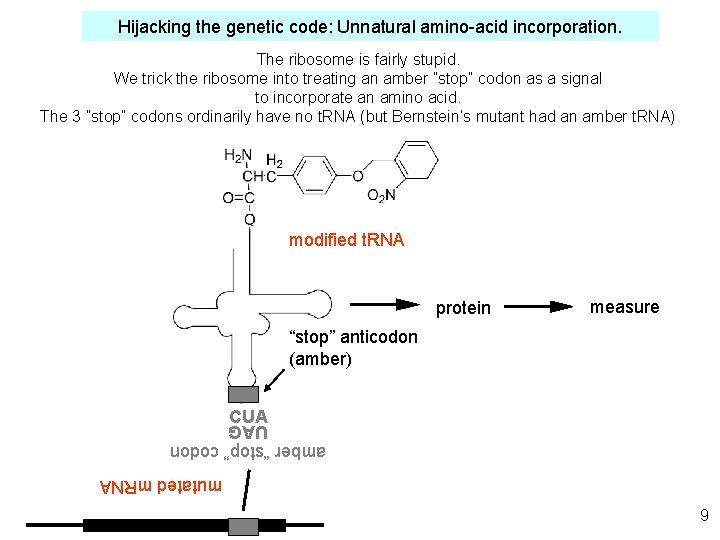
Hijacking the genetic code: Unnatural amino-acid incorporation. The ribosome is fairly stupid. We trick the ribosome into treating an amber ”stop” codon as a signal to incorporate an amino acid. The 3 ”stop” codons ordinarily have no t. RNA (but Bernstein’s mutant had an amber t. RNA) modified t. RNA protein measure “stop” anticodon (amber) CUA amber “stop” codon UAG mutated m. RNA 9

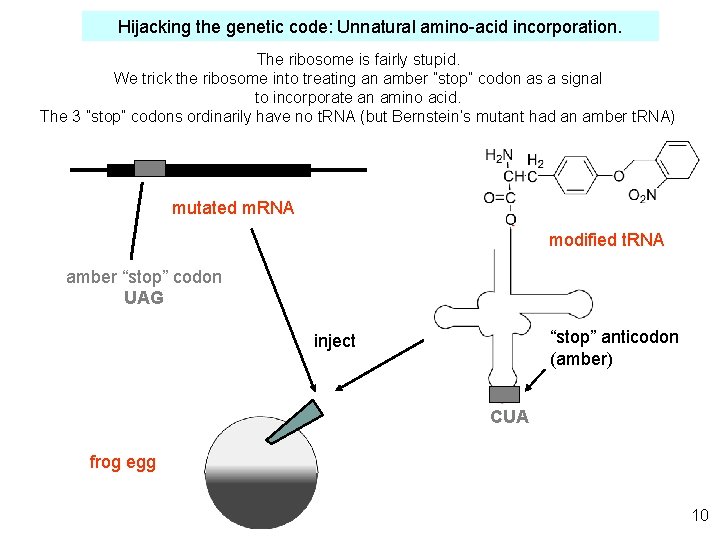
Hijacking the genetic code: Unnatural amino-acid incorporation. The ribosome is fairly stupid. We trick the ribosome into treating an amber ”stop” codon as a signal to incorporate an amino acid. The 3 ”stop” codons ordinarily have no t. RNA (but Bernstein’s mutant had an amber t. RNA) mutated m. RNA modified t. RNA amber “stop” codon UAG “stop” anticodon (amber) inject CUA frog egg 10

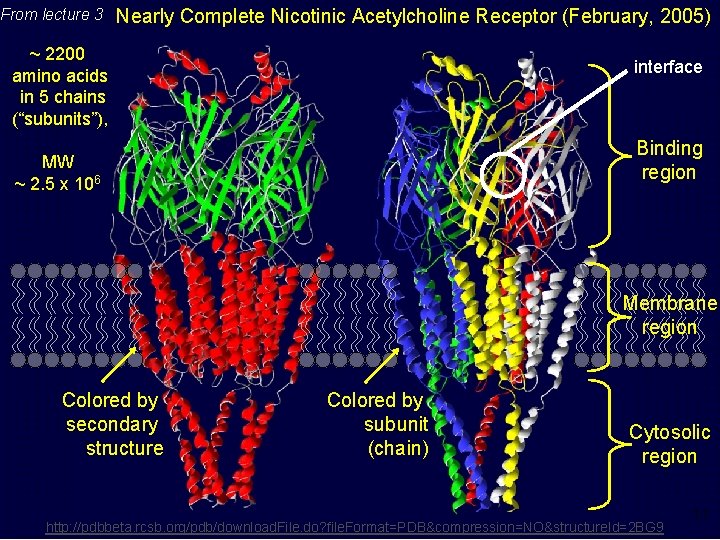
From lecture 3 Nearly Complete Nicotinic Acetylcholine Receptor (February, 2005) ~ 2200 amino acids in 5 chains (“subunits”), interface Binding region MW ~ 2. 5 x 106 Membrane region Colored by secondary structure Colored by subunit (chain) Cytosolic region http: //pdbbeta. rcsb. org/pdb/download. File. do? file. Format=PDB&compression=NO&structure. Id=2 BG 9 11

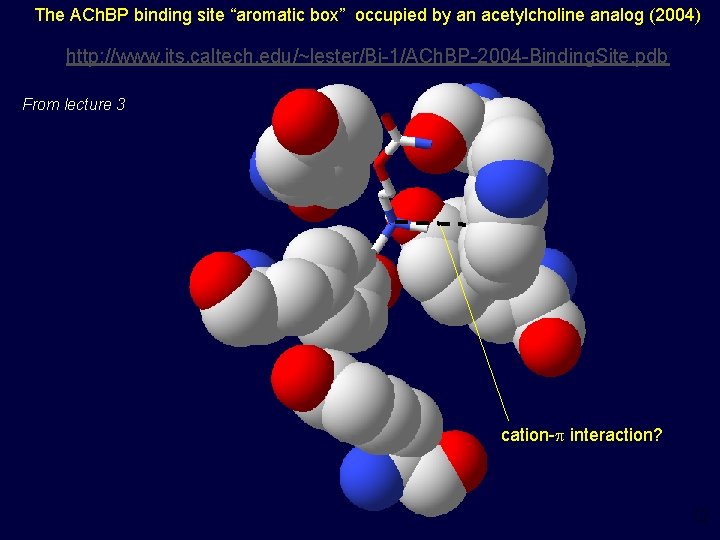
The ACh. BP binding site “aromatic box” occupied by an acetylcholine analog (2004) http: //www. its. caltech. edu/~lester/Bi-1/ACh. BP-2004 -Binding. Site. pdb From lecture 3 http: //www. its. caltech. edu/~lester/Bi-1 -2004/ACh. BP-2004 -Binding. Site. pdb cation-p interaction? 12

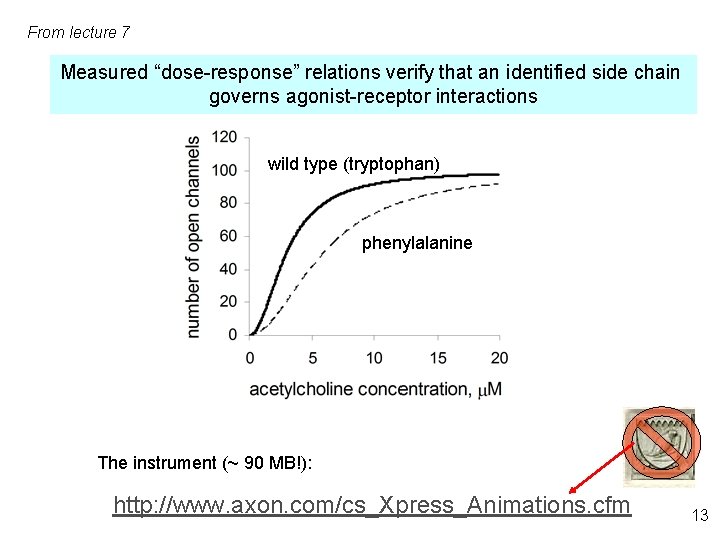
From lecture 7 Measured “dose-response” relations verify that an identified side chain governs agonist-receptor interactions wild type (tryptophan) phenylalanine The instrument (~ 90 MB!): http: //www. axon. com/cs_Xpress_Animations. cfm 13

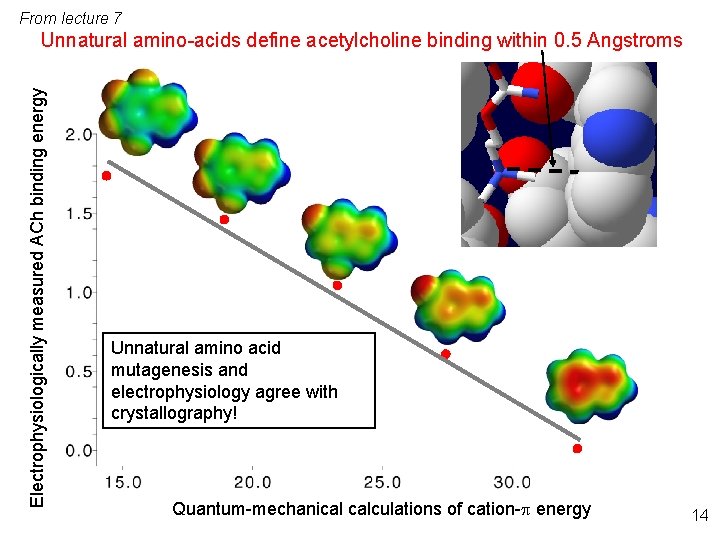
From lecture 7 Electrophysiologically measured ACh binding energy Unnatural amino-acids define acetylcholine binding within 0. 5 Angstroms Unnatural amino acid mutagenesis and electrophysiology agree with crystallography! Quantum-mechanical calculations of cation-p energy 14

B. Protein degradation is accomplished primarily by proteolytic enzymes The genome encodes hundreds of proteolytic enzymes. They vary in -- sequence specificity for the “cut” -- cellular expression -- organelle of expression 15

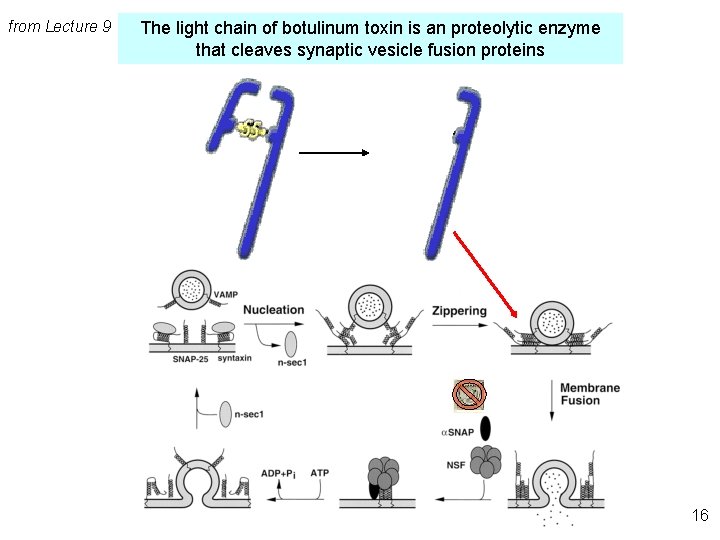
from Lecture 9 The light chain of botulinum toxin is an proteolytic enzyme that cleaves synaptic vesicle fusion proteins 16

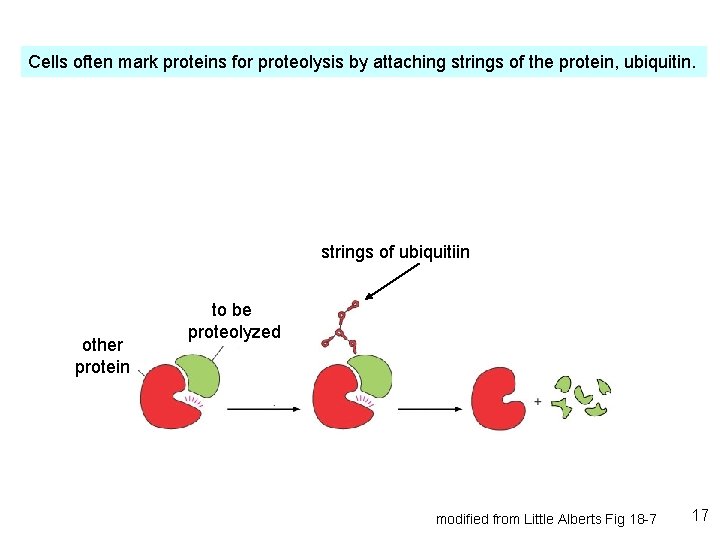
Cells often mark proteins for proteolysis by attaching strings of the protein, ubiquitin. strings of ubiquitiin other protein to be proteolyzed modified from Little Alberts Fig 18 -7 17

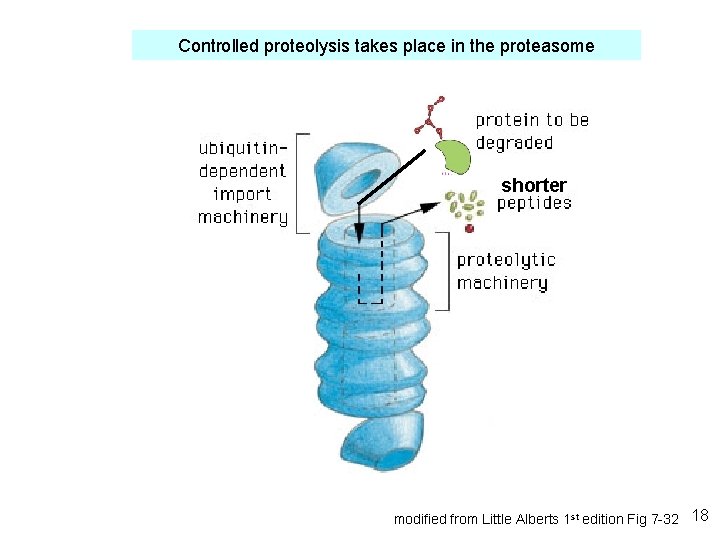
Controlled proteolysis takes place in the proteasome shorter modified from Little Alberts 1 st edition Fig 7 -32 18

Controlled, selective proteolysis is vital for healthy cells. . . Failed protein breakdown may help to cause some neurodegenerative diseases such as Huntington’s, Parkinson’s and Alzheimer’s Lectures 22, 26 below 19

Who pays for all this research? The US National Institutes of Health http: //maps. yahoo. com/maps_result? addr=&csz=Bethesda+MD&country=&new=1 20

Executive Branch 2006 NIH Budget $28. 4 B Department of Health and Human Services The US National Institutes of Health (NIH) 1. On-campus (“intramural”) research accounts for 10% 2. The majority of the budget is awarded as research grants (~ 9, 500) Bethesda MD, Just outside Washington DC Across the street: Walter Reed Army Hospital Uniformed Services University of the Health Sciences Howard Hughes Medical Institute Some FDA Research 21

National Institutes of Health: Institutes and Centers http: //www. nih. gov/icd/ 22

Monday 5/8. Dr. Michael Mc. Intosh will introduce psychiatric disease. Bi 1 “Drugs and the Brain” End of Lecture 18 23