Comparative analysis of host response transcriptomic and proteomic
- Slides: 1

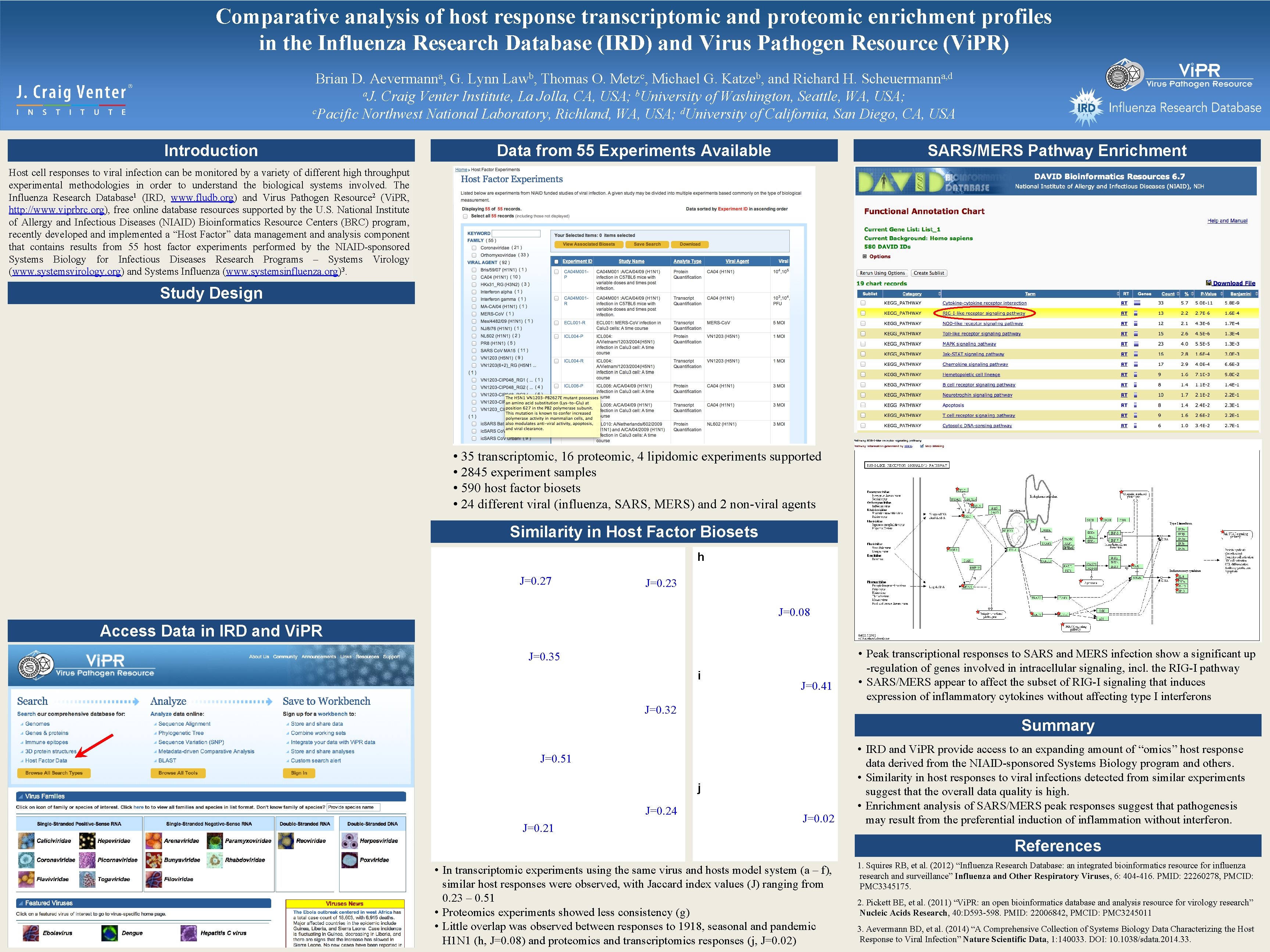
Comparative analysis of host response transcriptomic and proteomic enrichment profiles in the Influenza Research Database (IRD) and Virus Pathogen Resource (Vi. PR) Brian D. Aevermanna, G. Lynn Lawb, Thomas O. Metzc, Michael G. Katzeb, and Richard H. Scheuermanna, d a. J. Craig Venter Institute, La Jolla, CA, USA; b. University of Washington, Seattle, WA, USA; c. Pacific Northwest National Laboratory, Richland, WA, USA; d. University of California, San Diego, CA, USA Introduction Data from 55 Experiments Available SARS/MERS Pathway Enrichment Host cell responses to viral infection can be monitored by a variety of different high throughput experimental methodologies in order to understand the biological systems involved. The Influenza Research Database 1 (IRD, www. fludb. org) and Virus Pathogen Resource 2 (Vi. PR, http: //www. viprbrc. org), free online database resources supported by the U. S. National Institute of Allergy and Infectious Diseases (NIAID) Bioinformatics Resource Centers (BRC) program, recently developed and implemented a “Host Factor” data management and analysis component that contains results from 55 host factor experiments performed by the NIAID-sponsored Systems Biology for Infectious Diseases Research Programs – Systems Virology (www. systemsvirology. org) and Systems Influenza (www. systemsinfluenza. org)3. Study Design • 35 transcriptomic, 16 proteomic, 4 lipidomic experiments supported • 2845 experiment samples • 590 host factor biosets • 24 different viral (influenza, SARS, MERS) and 2 non-viral agents Similarity in Host Factor Biosets h J=0. 27 J=0. 23 J=0. 08 Access Data in IRD and Vi. PR J=0. 35 i J=0. 41 J=0. 32 Summary J=0. 51 j J=0. 24 J=0. 21 • Peak transcriptional responses to SARS and MERS infection show a significant up -regulation of genes involved in intracellular signaling, incl. the RIG-I pathway • SARS/MERS appear to affect the subset of RIG-I signaling that induces expression of inflammatory cytokines without affecting type I interferons J=0. 02 • IRD and Vi. PR provide access to an expanding amount of “omics” host response data derived from the NIAID-sponsored Systems Biology program and others. • Similarity in host responses to viral infections detected from similar experiments suggest that the overall data quality is high. • Enrichment analysis of SARS/MERS peak responses suggest that pathogenesis may result from the preferential induction of inflammation without interferon. References • In transcriptomic experiments using the same virus and hosts model system (a – f), similar host responses were observed, with Jaccard index values (J) ranging from 0. 23 – 0. 51 • Proteomics experiments showed less consistency (g) • Little overlap was observed between responses to 1918, seasonal and pandemic H 1 N 1 (h, J=0. 08) and proteomics and transcriptomics responses (j, J=0. 02) 1. Squires RB, et al. (2012) “Influenza Research Database: an integrated bioinformatics resource for influenza research and surveillance” Influenza and Other Respiratory Viruses, 6: 404 -416. PMID: 22260278, PMCID: PMC 3345175. 2. Pickett BE, et al. (2011) “Vi. PR: an open bioinformatics database and analysis resource for virology research” Nucleic Acids Research, 40: D 593 -598. PMID: 22006842, PMCID: PMC 3245011 3. Aevermann BD, et al. (2014) “A Comprehensive Collection of Systems Biology Data Characterizing the Host Response to Viral Infection” Nature Scientific Data, 1: 140033. DOI: 10. 1038/sdata. 2014. 33.