Class 37 Secret of Life CS 200 Computer



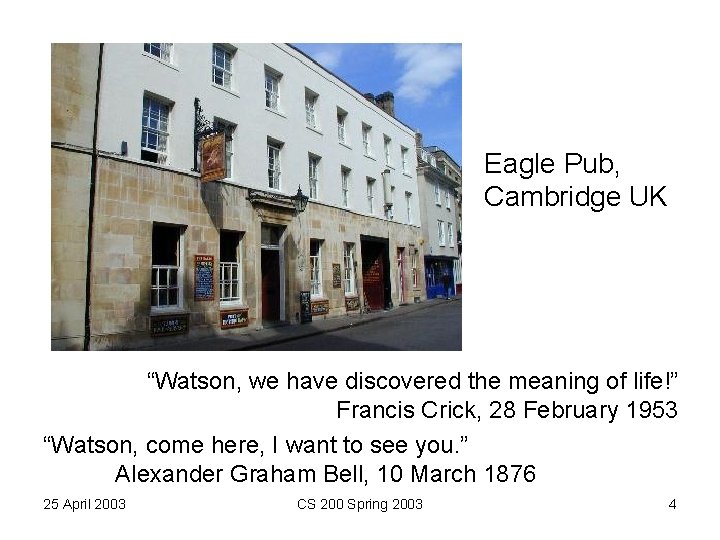














- Slides: 33

Class 37: Secret of Life CS 200: Computer Science David Evans University of Virginia 25 April 2003 CS 200 Spring 2003 http: //www. cs. virginia. edu/~evans Computer Science

From Lecture 15: Liberal Arts Quadrivium Trivium • Grammar: study of meaning in written BNF replacement rules for describing languages, expression rules of evaluation for meaning • Rhetoric: comprehension of and written discourse verbal Your. Not PS 8 web sites arebetween a discourse yet… Interfaces components, program and user between user and server. • Logic: argumentative discourse discovering truth for Rules of evaluation, if, recursive definitions Learned to count Not much yet…in Lambda Calculus wait until April • Arithmetic: understanding numbers • Geometry: quantification of space • Music: number in time • Astronomy 25 April 2003 Curves as procedures, fractals Yes, even if we can’t figure out how to play “Hey Jude!” Yes: Neil de. Grasse Tyson says so CS 200 Spring 2003 2

th 50 Today is the anniversary of announcement of the most important scientific discovery of th the 20 century! 25 April 2003 CS 200 Spring 2003 3
Eagle Pub, Cambridge UK “Watson, we have discovered the meaning of life!” Francis Crick, 28 February 1953 “Watson, come here, I want to see you. ” Alexander Graham Bell, 10 March 1876 25 April 2003 CS 200 Spring 2003 4

Molecular Structure of Nucleic Acids, “A Structure for Deoxyribose Nucleic Acid”, Nature 25 April 1953 It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material. http: //www. nature. com/genomics/human/watson-crick/watson_crick. pdf 25 April 2003

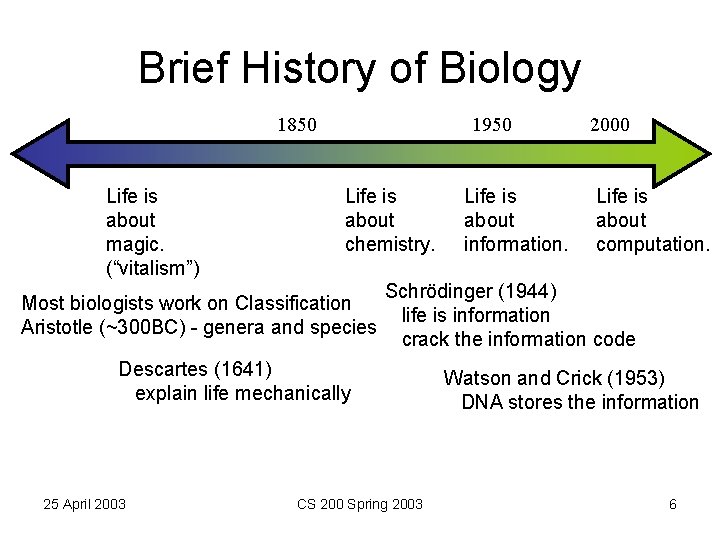
Brief History of Biology 1950 1850 Life is about magic. (“vitalism”) Life is about chemistry. Most biologists work on Classification Aristotle (~300 BC) - genera and species Life is about computation. Schrödinger (1944) life is information crack the information code Descartes (1641) explain life mechanically 25 April 2003 Life is about information. 2000 CS 200 Spring 2003 Watson and Crick (1953) DNA stores the information 6

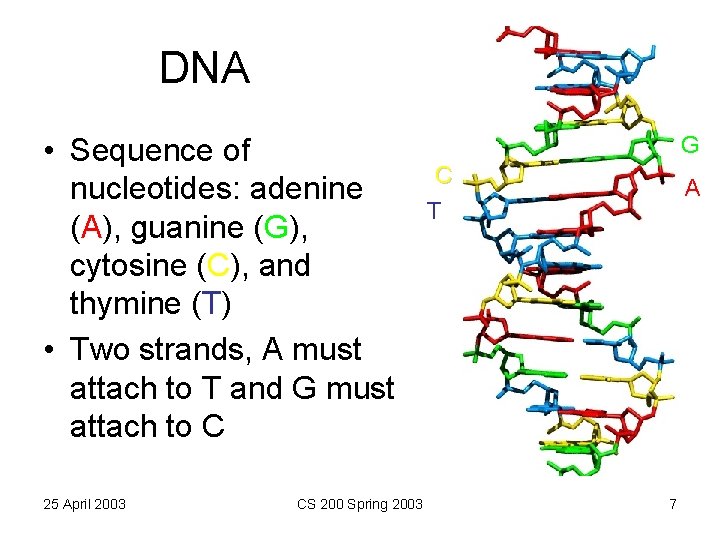
DNA • Sequence of nucleotides: adenine (A), guanine (G), cytosine (C), and thymine (T) • Two strands, A must attach to T and G must attach to C 25 April 2003 CS 200 Spring 2003 G C T A 7

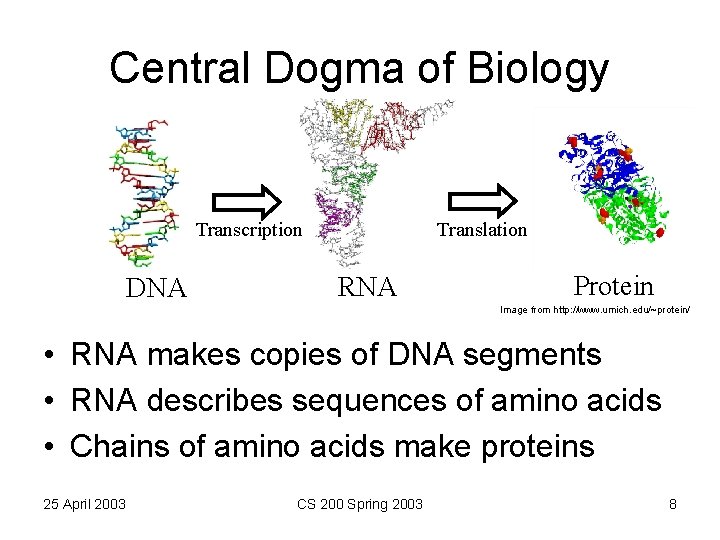
Central Dogma of Biology Translation Transcription DNA RNA Protein Image from http: //www. umich. edu/~protein/ • RNA makes copies of DNA segments • RNA describes sequences of amino acids • Chains of amino acids make proteins 25 April 2003 CS 200 Spring 2003 8

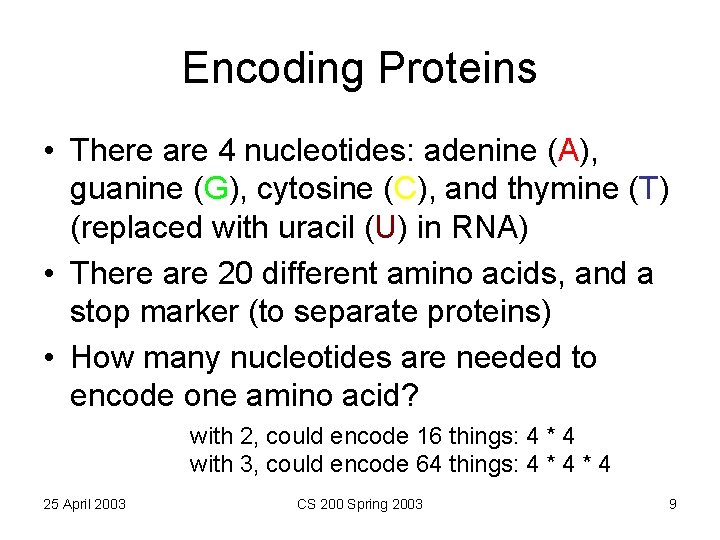
Encoding Proteins • There are 4 nucleotides: adenine (A), guanine (G), cytosine (C), and thymine (T) (replaced with uracil (U) in RNA) • There are 20 different amino acids, and a stop marker (to separate proteins) • How many nucleotides are needed to encode one amino acid? with 2, could encode 16 things: 4 * 4 with 3, could encode 64 things: 4 * 4 25 April 2003 CS 200 Spring 2003 9

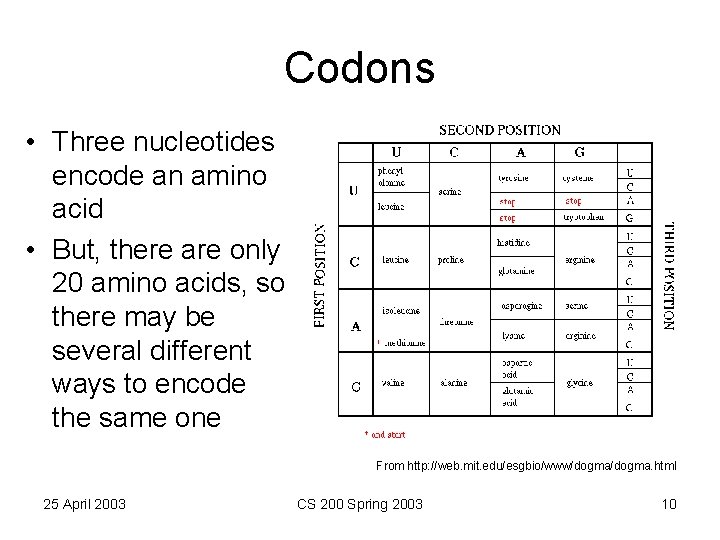
Codons • Three nucleotides encode an amino acid • But, there are only 20 amino acids, so there may be several different ways to encode the same one From http: //web. mit. edu/esgbio/www/dogma. html 25 April 2003 CS 200 Spring 2003 10

How Big is the Make-a-Human Program? • 3 Billion Base Pairs – Each nucleotide is 2 bits (4 possibilities) – 3 B pairs * 1 byte/4 pairs = 750 MB • Every sequence of 3 base pairs one of 20 amino acids (or stop codon) – 21 possible codons, but 43 = 64 possible – So, really only 750 MB * (21/64) ~ 250 MB 25 April 2003 CS 200 Spring 2003 11

1 CD ~ 650 MB 25 April 2003 CS 200 Spring 2003 12

People are almost all the Same • Genetic code for 2 humans differs in only 2. 1 million bases – 4 million bits = 0. 5 MB • How big is 0. 5 MB? – 1/3 of a floppy disk – ~22 times the size of the PS 6 adventure game code 25 April 2003 CS 200 Spring 2003 13

Is DNA Really a Programming Language? 25 April 2003 CS 200 Spring 2003 14

Stuff Programming Languages are Made Of • Primitives codons (sequence of 3 nucleotides that encodes a protein) • Means of Combination ? ? Morphogenesis? Not well understood (by anyone). This is where most of the expressiveness comes from! • Means of Abstraction DNA itself – separate proteins from their encoding Genes – group DNA by function (sort of) Chromosomes – package Genes together Organisms – packages for reproducing Genes 25 April 2003 CS 200 Spring 2003 15

My Research Group • Build robust, survivable systems from unreliable components – Learn from biological systems that do this • Cell-Based Programming Model – Genes turn on and off state changes – Emit different chemicals depending on state, sense chemicals in surroundings – Cells can divide asymmetrically – Lots of simplifications: not simulating reality 25 April 2003 CS 200 Spring 2003 16

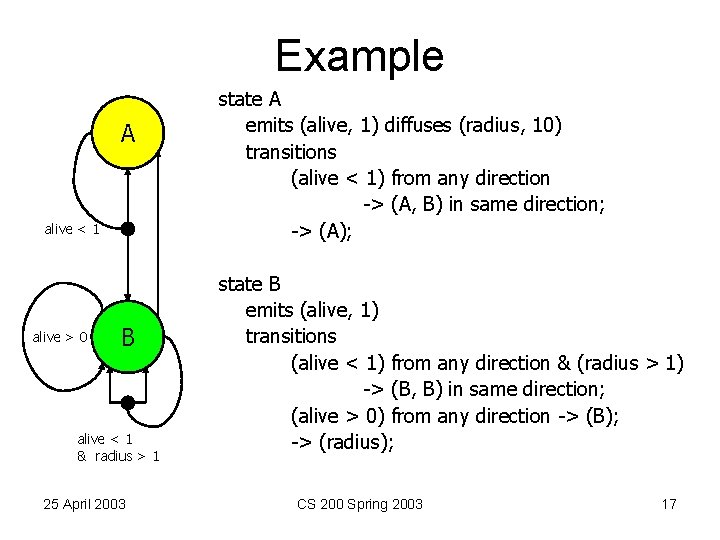
Example A alive < 1 alive > 0 B alive < 1 & radius > 1 25 April 2003 state A emits (alive, 1) diffuses (radius, 10) transitions (alive < 1) from any direction -> (A, B) in same direction; -> (A); state B emits (alive, 1) transitions (alive < 1) from any direction & (radius > 1) -> (B, B) in same direction; (alive > 0) from any direction -> (B); -> (radius); CS 200 Spring 2003 17

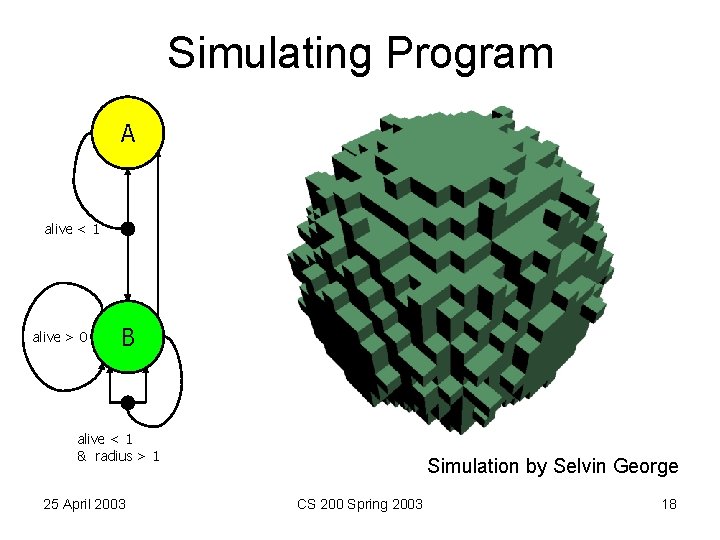
Simulating Program A alive < 1 alive > 0 B alive < 1 & radius > 1 25 April 2003 Simulation by Selvin George CS 200 Spring 2003 18

Simulation by Selvin George 25 April 2003 CS 200 Spring 2003 19

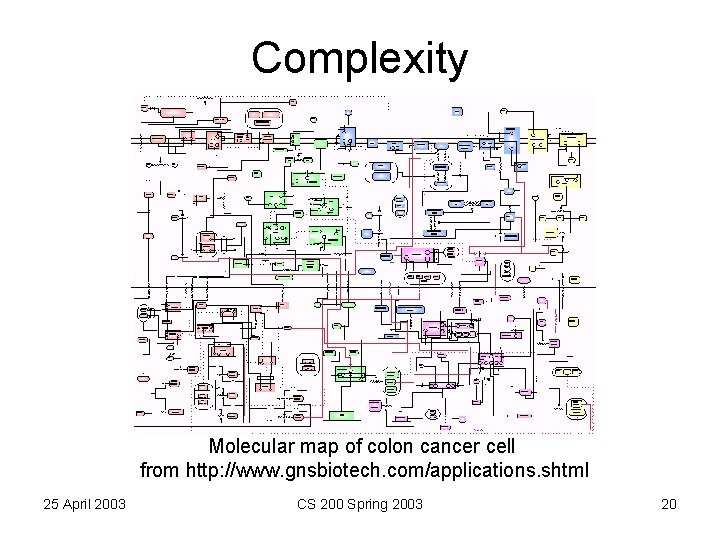
Complexity Molecular map of colon cancer cell from http: //www. gnsbiotech. com/applications. shtml 25 April 2003 CS 200 Spring 2003 20

Computing with DNA Leonard Adleman (Mathematical Consultant for Sneakers), 1995 25 April 2003 CS 200 Spring 2003 21

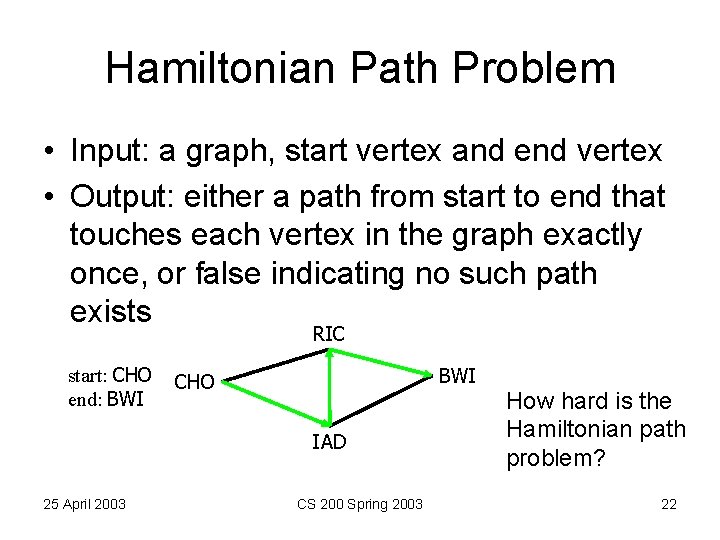
Hamiltonian Path Problem • Input: a graph, start vertex and end vertex • Output: either a path from start to end that touches each vertex in the graph exactly once, or false indicating no such path exists RIC start: CHO end: BWI CHO IAD 25 April 2003 CS 200 Spring 2003 How hard is the Hamiltonian path problem? 22

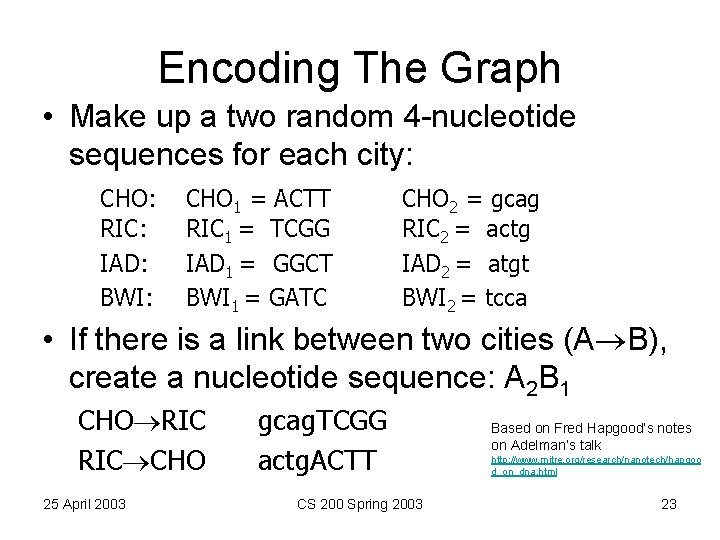
Encoding The Graph • Make up a two random 4 -nucleotide sequences for each city: CHO: RIC: IAD: BWI: CHO 1 = ACTT RIC 1 = TCGG IAD 1 = GGCT BWI 1 = GATC CHO 2 = gcag RIC 2 = actg IAD 2 = atgt BWI 2 = tcca • If there is a link between two cities (A B), create a nucleotide sequence: A 2 B 1 CHO RIC CHO 25 April 2003 gcag. TCGG actg. ACTT CS 200 Spring 2003 Based on Fred Hapgood’s notes on Adelman’s talk http: //www. mitre. org/research/nanotech/hapgoo d_on_dna. html 23

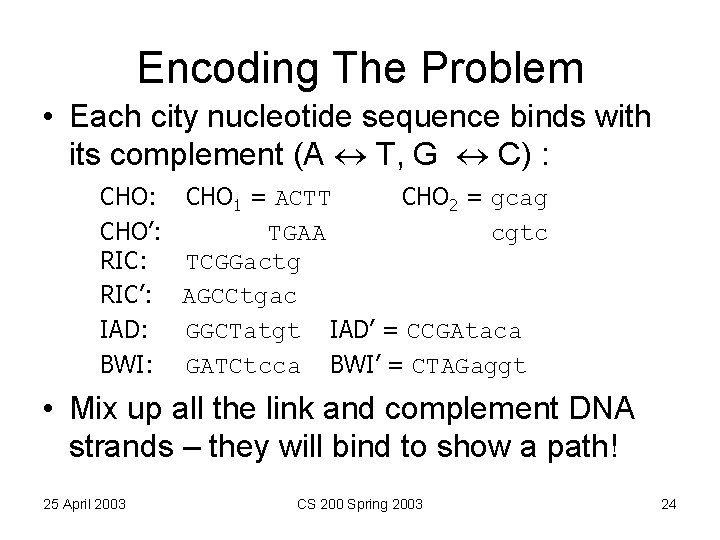
Encoding The Problem • Each city nucleotide sequence binds with its complement (A T, G C) : CHO 1 = ACTT CHO 2 = gcag CHO’: TGAA cgtc RIC: TCGGactg RIC’: AGCCtgac IAD: GGCTatgt IAD’ = CCGAtaca BWI: GATCtcca BWI’ = CTAGaggt • Mix up all the link and complement DNA strands – they will bind to show a path! 25 April 2003 CS 200 Spring 2003 24

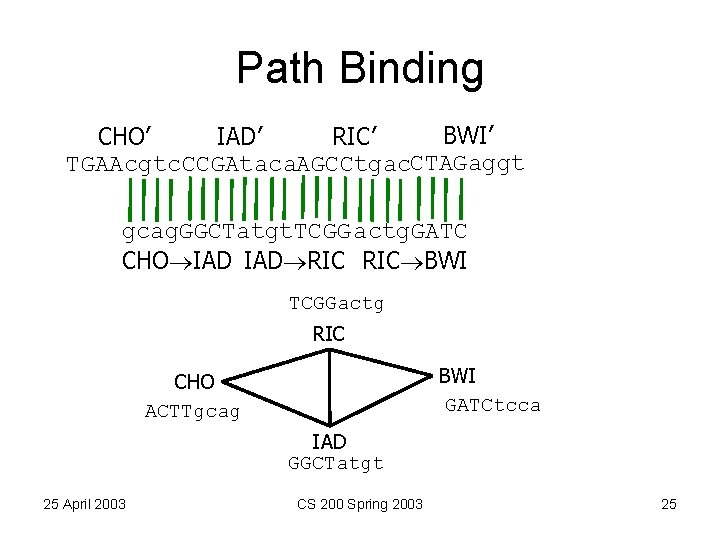
Path Binding BWI’ RIC’ IAD’ CHO’ TGAAcgtc. CCGAtaca. AGCCtgac. CTAGaggt gcag. GGCTatgt. TCGG actg. GATC CHO IAD RIC BWI TCGGactg RIC BWI GATCtcca CHO ACTTgcag IAD GGCTatgt 25 April 2003 CS 200 Spring 2003 25

Getting the Solution • Extract DNA strands starting with CHO and ending with BWI – Easy way is to remove all strands that do not start with CHO, and then remove all strands that do not end with BWI • Measure remaining strands to find ones with the right weight (7 * 8 nucleotides) • Read the sequence from one of these strands 25 April 2003 CS 200 Spring 2003 26

Why don’t we use DNA computers? • Speed: shaking up the DNA strands does 1014 operations per second ($400 M supercomputer does 1010) • Memory: we can store information in DNA at 1 bit per cubic nanometer • How much DNA would you need? – Volume of DNA needed grows exponentially with input size – To solve ~45 vertices, you need ~20 M gallons 25 April 2003 CS 200 Spring 2003 27

DNA-Enhanced PC 25 April 2003 CS 200 Spring 2003 28

Biology is (becoming) a subfield of Computer Science • Biological mechanisms are mostly understood (proteomics still has a way to go) • What is not understood is how those are combined to create meaning 25 April 2003 CS 200 Spring 2003 29

PS 8 • Before 10: 55 am Monday: – Submit a zip file of all your code using a form linked from the CS 200 web site – If you want to use a few Power. Point slides in your presentation, you may submit those also • You only have 3 or 5 minutes: use them wisely – Figure out beforehand what you will do – Recommend: one team member drive web browser, one (or two) talk – Talk about what users should know about your website, not about how you built it (unless there is something especially interesting) 25 April 2003 CS 200 Spring 2003 30

Mc. Intire Symposium Talk: Daniel Kahneman (Psychologist, Nobel Prize in Economics) • When you are 99% sure, how often are you actually right? – 85 -90% of the time – Some of you will get a sticker on your Exam 2 that will make you 99. 5% sure of the lowest grade you could receive in CS 200 (the 0. 5% is since you still need to do PS 8 well) • Humans are overly optimistic and excessively risk averse – No risk in taking the final: it cannot lower your grade – You should be optimistic that it can help your grade 25 April 2003 CS 200 Spring 2003 31

Final • Out Monday, due Monday, May 5 (4: 55 pm) • You have 8 days, but should not spend more than 4 hours on the exam • Will include: – A small programming problem (like a PS) – Some questions about computability and complexity 25 April 2003 CS 200 Spring 2003 32

Graduation Photo 25 April 2003 CS 200 Spring 2003 33
 300+200+200+200
300+200+200+200 The secret life of walter mitty quotes
The secret life of walter mitty quotes Life of an inflatable secret weapon
Life of an inflatable secret weapon Secret life of bees characterization
Secret life of bees characterization Feminism in the secret life of bees
Feminism in the secret life of bees Allusions in secret life of bees
Allusions in secret life of bees Symbolism literary device
Symbolism literary device 200+200+100+100
200+200+100+100 200+400+600+800
200+400+600+800 300 + 200 + 200
300 + 200 + 200 200+200+100
200+200+100 100 + 100 200
100 + 100 200 100 200 300
100 200 300 Kth print
Kth print Are there ethical reasons to try to crack secret codes?
Are there ethical reasons to try to crack secret codes? Listerine trade secret case
Listerine trade secret case The secrets of successful strategy execution
The secrets of successful strategy execution The secret lives of sgt john wilson timeline
The secret lives of sgt john wilson timeline Secret garden setting
Secret garden setting Judge the crucible
Judge the crucible Secret class129
Secret class129 The secret heart by robert p. tristram coffin
The secret heart by robert p. tristram coffin My secret place poem
My secret place poem A department store decides to use secret shoppers
A department store decides to use secret shoppers Setting of the secret garden
Setting of the secret garden Agence famille sncf pass carmillon
Agence famille sncf pass carmillon Simple secret key distribution
Simple secret key distribution Passive attack
Passive attack Jardin secret rumilly
Jardin secret rumilly Difference between copyright and patent
Difference between copyright and patent Top secret telegram
Top secret telegram Google hacking index of
Google hacking index of Cryptography definition
Cryptography definition Simple secret key distribution
Simple secret key distribution