Vespa Versatile Simulation Pulses and Analysis Created via


- Slides: 12

Vespa – Versatile Simulation, Pulses and Analysis • Created via an NIH software support/maintenance grant • Derived from three existing applications on the ‘sneakernet’ • Mat. Pulse – Mat. Lab • GAMMA – C++ • SITools/FITT – IDL/C/Fortran URL: http: //scion. duhs. duke. edu/vespa

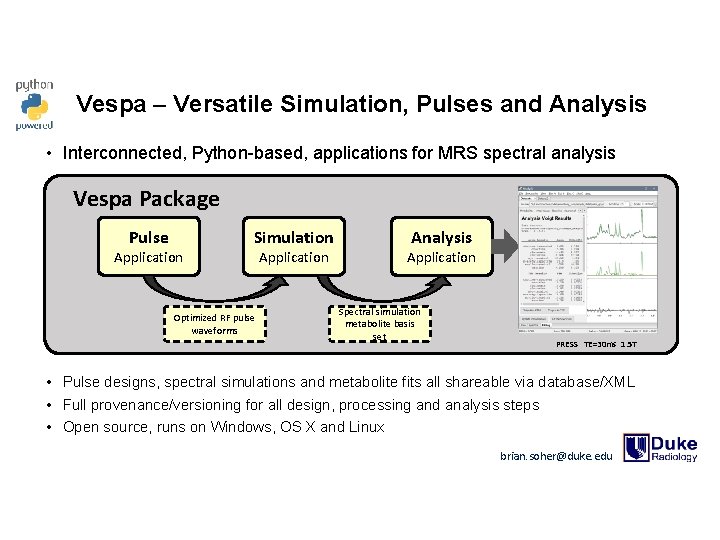
Vespa – Versatile Simulation, Pulses and Analysis • Interconnected, Python-based, applications for MRS spectral analysis Vespa Package Pulse Application Simulation Optimized RF pulse waveforms Application Analysis Application Spectral simulation metabolite basis set PRESS TE=30 ms 1. 5 T • Pulse designs, spectral simulations and metabolite fits all shareable via database/XML • Full provenance/versioning for all design, processing and analysis steps • Open source, runs on Windows, OS X and Linux brian. soher@duke. edu

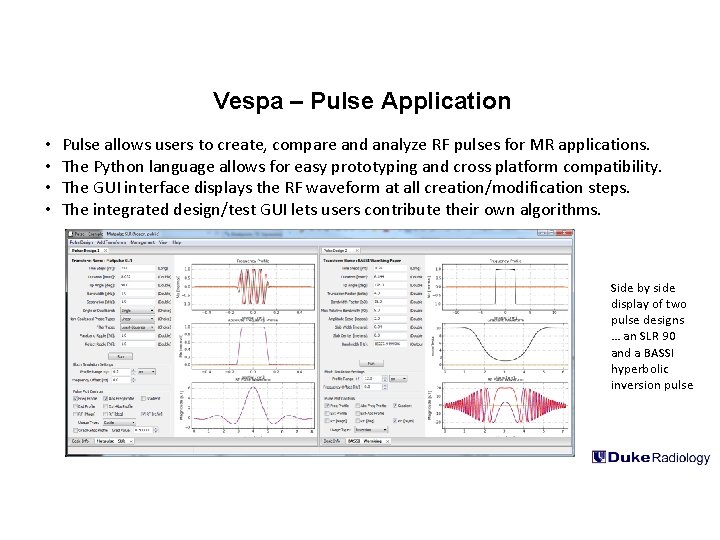
Vespa – Pulse Application • • Pulse allows users to create, compare and analyze RF pulses for MR applications. The Python language allows for easy prototyping and cross platform compatibility. The GUI interface displays the RF waveform at all creation/modification steps. The integrated design/test GUI lets users contribute their own algorithms. Side by side display of two pulse designs … an SLR 90 and a BASSI hyperbolic inversion pulse

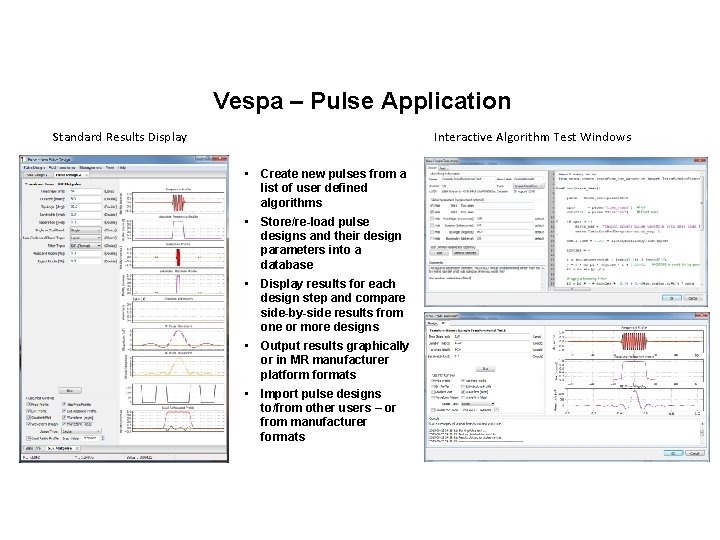
Vespa – Pulse Application Standard Results Display Interactive Algorithm Test Windows • Create new pulses from a list of user defined algorithms • Store/re-load pulse designs and their design parameters into a database • Display results for each design step and compare side-by-side results from one or more designs • Output results graphically or in MR manufacturer platformats • Import pulse designs to/from other users – or from manufacturer formats

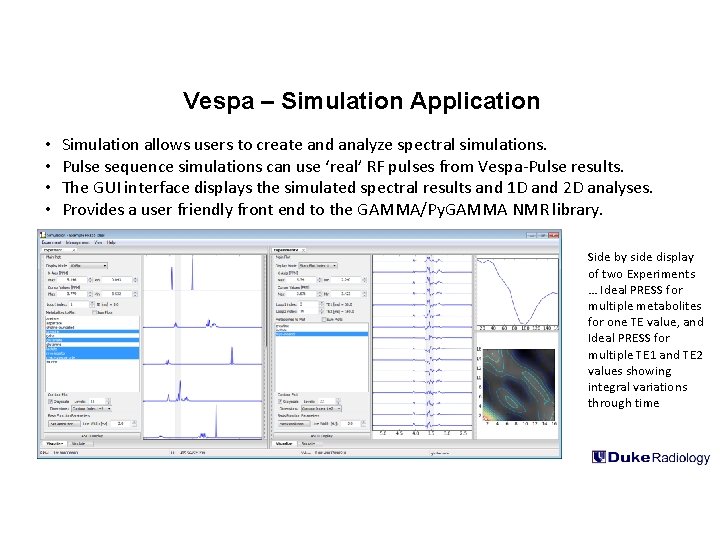
Vespa – Simulation Application • • Simulation allows users to create and analyze spectral simulations. Pulse sequence simulations can use ‘real’ RF pulses from Vespa-Pulse results. The GUI interface displays the simulated spectral results and 1 D and 2 D analyses. Provides a user friendly front end to the GAMMA/Py. GAMMA NMR library. Side by side display of two Experiments … Ideal PRESS for multiple metabolites for one TE value, and Ideal PRESS for multiple TE 1 and TE 2 values showing integral variations through time

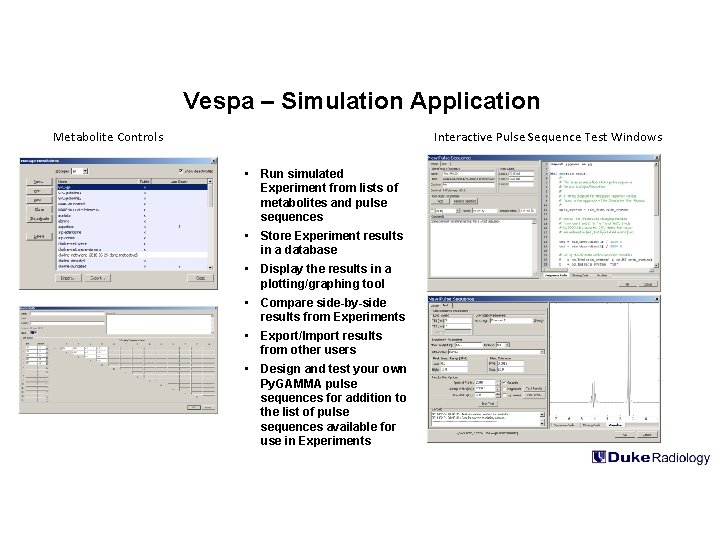
Vespa – Simulation Application Metabolite Controls Interactive Pulse Sequence Test Windows • Run simulated Experiment from lists of metabolites and pulse sequences • Store Experiment results in a database • Display the results in a plotting/graphing tool • Compare side-by-side results from Experiments • Export/Import results from other users • Design and test your own Py. GAMMA pulse sequences for addition to the list of pulse sequences available for use in Experiments

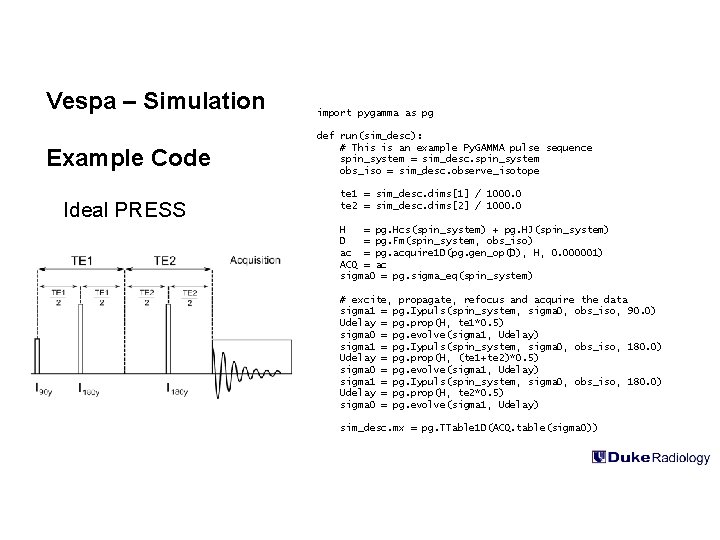
Vespa – Simulation import pygamma as pg Example Code def run(sim_desc): # This is an example Py. GAMMA pulse sequence spin_system = sim_desc. spin_system obs_iso = sim_desc. observe_isotope Ideal PRESS te 1 = sim_desc. dims[1] / 1000. 0 te 2 = sim_desc. dims[2] / 1000. 0 H = pg. Hcs(spin_system) + pg. HJ(spin_system) D = pg. Fm(spin_system, obs_iso) ac = pg. acquire 1 D(pg. gen_op(D), H, 0. 000001) ACQ = ac sigma 0 = pg. sigma_eq(spin_system) # excite, propagate, refocus and acquire the data sigma 1 = pg. Iypuls(spin_system, sigma 0, obs_iso, 90. 0) Udelay = pg. prop(H, te 1*0. 5) sigma 0 = pg. evolve(sigma 1, Udelay) sigma 1 = pg. Iypuls(spin_system, sigma 0, obs_iso, 180. 0) Udelay = pg. prop(H, (te 1+te 2)*0. 5) sigma 0 = pg. evolve(sigma 1, Udelay) sigma 1 = pg. Iypuls(spin_system, sigma 0, obs_iso, 180. 0) Udelay = pg. prop(H, te 2*0. 5) sigma 0 = pg. evolve(sigma 1, Udelay) sim_desc. mx = pg. TTable 1 D(ACQ. table(sigma 0))

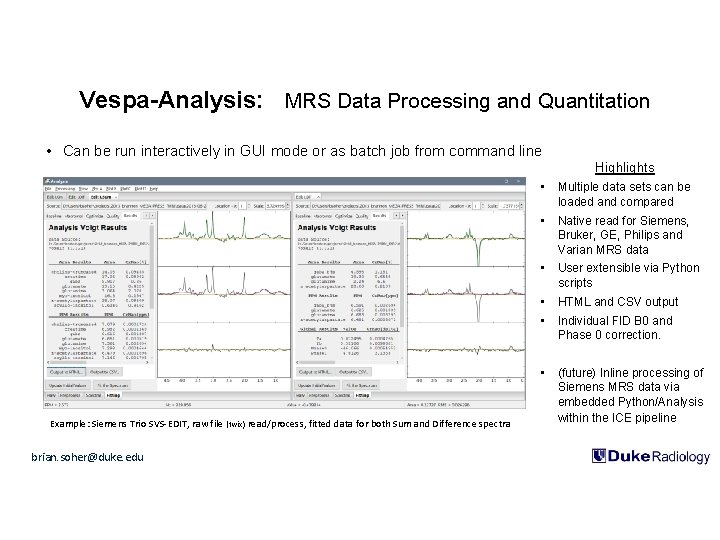
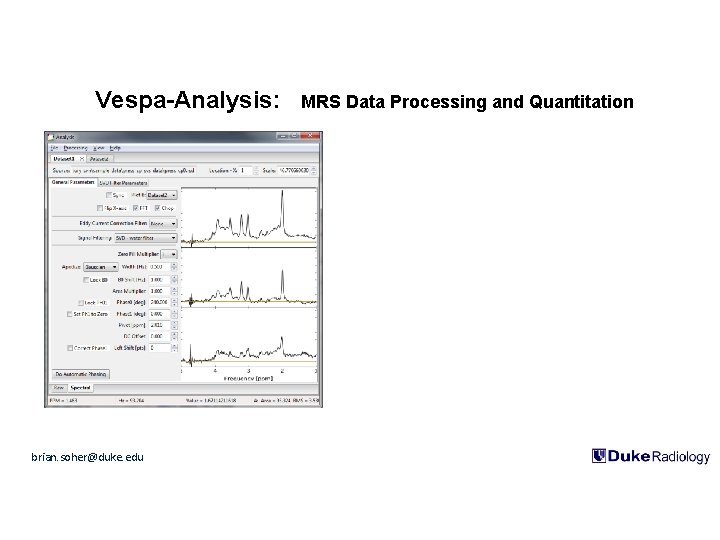
Vespa-Analysis: MRS Data Processing and Quantitation • Can be run interactively in GUI mode or as batch job from command line Highlights Example: Siemens Trio SVS-EDIT, raw file (twix) read/process, fitted data for both Sum and Difference spectra brian. soher@duke. edu • Multiple data sets can be loaded and compared • Native read for Siemens, Bruker, GE, Philips and Varian MRS data • User extensible via Python scripts • HTML and CSV output • Individual FID B 0 and Phase 0 correction. • (future) Inline processing of Siemens MRS data via embedded Python/Analysis within the ICE pipeline

Vespa-Analysis: brian. soher@duke. edu MRS Data Processing and Quantitation

The Future and my Wish List • Inline processing of Siemens MRS data via embedded Python/Analysis within the ICE pipeline • Output to bitmap image of results • Standardized semi-LASER sequence and processing on Siemens and Philips • Want to wrap results into DICOM Structured Report • More centralized processing location – Syngo Via. Frontier server? • Also able to embed Python code • Ni. Pype – Neuroimaging in Python workflow package

Thank you!
