Traditional diagnostic testing vs Whole Exome Sequencing in
- Slides: 1

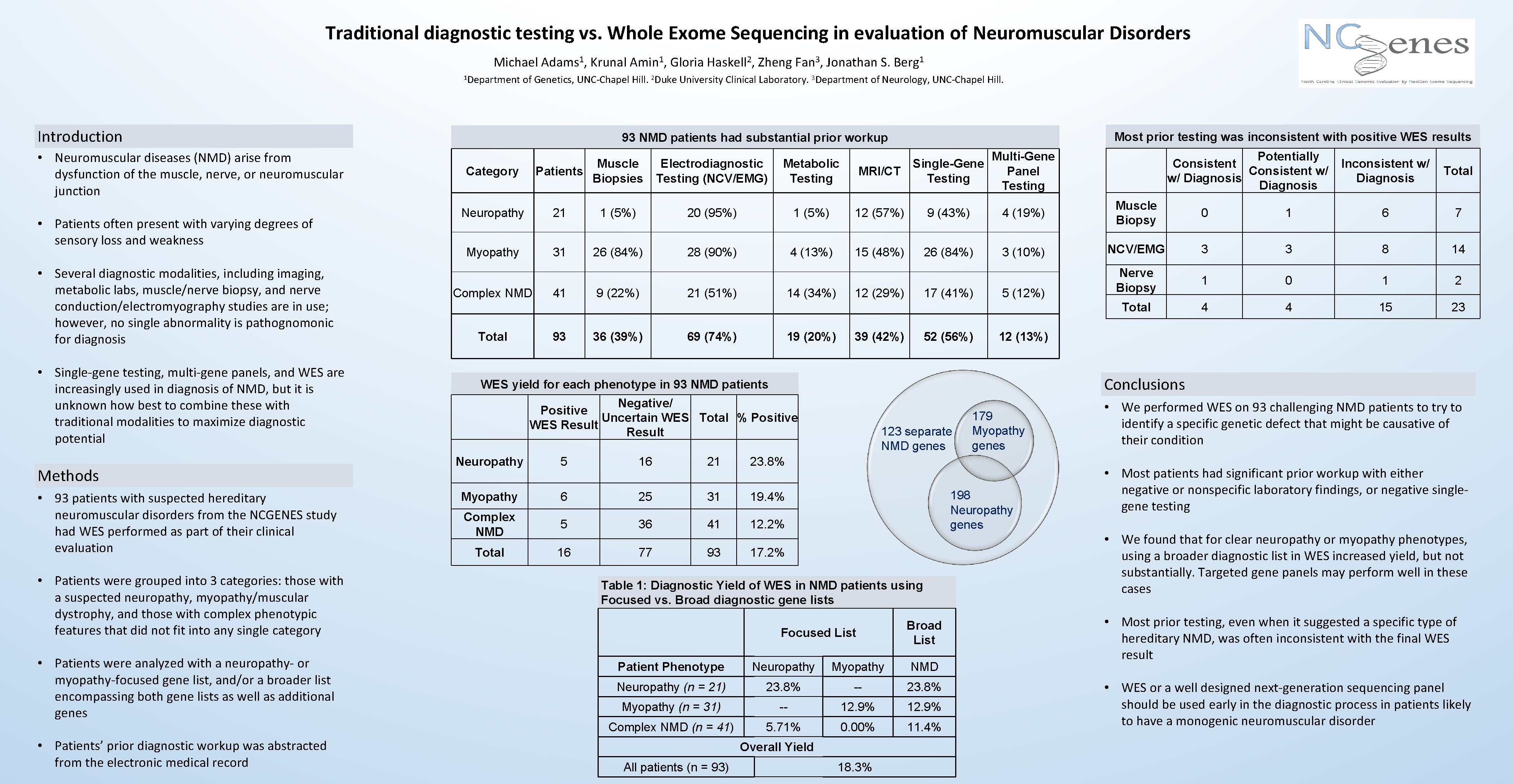
Traditional diagnostic testing vs. Whole Exome Sequencing in evaluation of Neuromuscular Disorders Michael Adams 1, Krunal Amin 1, Gloria Haskell 2, Zheng Fan 3, Jonathan S. Berg 1 1 Department of Genetics, UNC-Chapel Hill. 2 Duke University Clinical Laboratory. 3 Department of Neurology, UNC-Chapel Hill. Introduction • Neuromuscular diseases (NMD) arise from dysfunction of the muscle, nerve, or neuromuscular junction • Patients often present with varying degrees of sensory loss and weakness • Several diagnostic modalities, including imaging, metabolic labs, muscle/nerve biopsy, and nerve conduction/electromyography studies are in use; however, no single abnormality is pathognomonic for diagnosis • Single-gene testing, multi-gene panels, and WES are increasingly used in diagnosis of NMD, but it is unknown how best to combine these with traditional modalities to maximize diagnostic potential Methods • 93 patients with suspected hereditary neuromuscular disorders from the NCGENES study had WES performed as part of their clinical evaluation • Patients were grouped into 3 categories: those with a suspected neuropathy, myopathy/muscular dystrophy, and those with complex phenotypic features that did not fit into any single category • Patients were analyzed with a neuropathy- or myopathy-focused gene list, and/or a broader list encompassing both gene lists as well as additional genes • Patients’ prior diagnostic workup was abstracted from the electronic medical record Most prior testing was inconsistent with positive WES results 93 NMD patients had substantial prior workup Muscle Patients Biopsies Category Electrodiagnostic Testing (NCV/EMG) Metabolic Testing MRI/CT Potentially Consistent w/ w/ Diagnosis Multi-Gene Single-Gene Panel Testing Inconsistent w/ Diagnosis Total Neuropathy 21 1 (5%) 20 (95%) 1 (5%) 12 (57%) 9 (43%) 4 (19%) Muscle Biopsy Myopathy 31 26 (84%) 28 (90%) 4 (13%) 15 (48%) 26 (84%) 3 (10%) NCV/EMG 3 3 8 14 5 (12%) Nerve Biopsy 1 0 1 2 Total 4 4 15 23 Complex NMD Total 41 93 9 (22%) 36 (39%) 21 (51%) 14 (34%) 69 (74%) 19 (20%) 12 (29%) 17 (41%) 39 (42%) 52 (56%) 1 6 7 12 (13%) Conclusions WES yield for each phenotype in 93 NMD patients Negative/ Positive Uncertain WES Total % Positive WES Result Neuropathy 5 16 21 23. 8% Myopathy 6 25 31 19. 4% Complex NMD 5 36 41 12. 2% Total 16 77 93 17. 2% 123 separate NMD genes 179 Myopathy genes 198 Neuropathy genes Table 1: Diagnostic Yield of WES in NMD patients using Focused vs. Broad diagnostic gene lists Focused List Broad List Patient Phenotype Neuropathy Myopathy NMD Neuropathy (n = 21) 23. 8% -- 23. 8% Myopathy (n = 31) -- 12. 9% Complex NMD (n = 41) 5. 71% 0. 00% 11. 4% Overall Yield All patients (n = 93) 0 18. 3% • We performed WES on 93 challenging NMD patients to try to identify a specific genetic defect that might be causative of their condition • Most patients had significant prior workup with either negative or nonspecific laboratory findings, or negative singlegene testing • We found that for clear neuropathy or myopathy phenotypes, using a broader diagnostic list in WES increased yield, but not substantially. Targeted gene panels may perform well in these cases • Most prior testing, even when it suggested a specific type of hereditary NMD, was often inconsistent with the final WES result • WES or a well designed next-generation sequencing panel should be used early in the diagnostic process in patients likely to have a monogenic neuromuscular disorder