MEGAN analysis of metagenomic data Daniel H Huson



- Slides: 17

MEGAN analysis of metagenomic data Daniel H. Huson, Alexander F. Auch, Ji Qi, et al. Genome Res. 2007

Early metagenomic � Known phylogenetic markers and subsequent sequencing of clones � Analysis of paired-end reads � Complete sequences of environmental fosmid and BAC clones Rough annotation of the metabolic capacity � Environmental assemblies Distinguish between discrete species and population of closely related biotypes � Problem of using proven phylogenetic markers(ribosomal genes, coding sequences) � Slow-evolving genes : distinguishing between species at large evolutionary distances

What is MEGAN? � Metagenome Analyzer (MEGAN) � Free software. � Deviates from the analytical pattern of previous � Built on the statistical analysis of comparing random sequence intervals with unspecified phylogenetic properties against databases � Depends on the related sequences in the databases � Providing filter to adjust the level of stringency later to an appropriate level � Laptop analysis � Comparing � Graphical result (BLAST)-> laptop (MEGAN) and statistical output

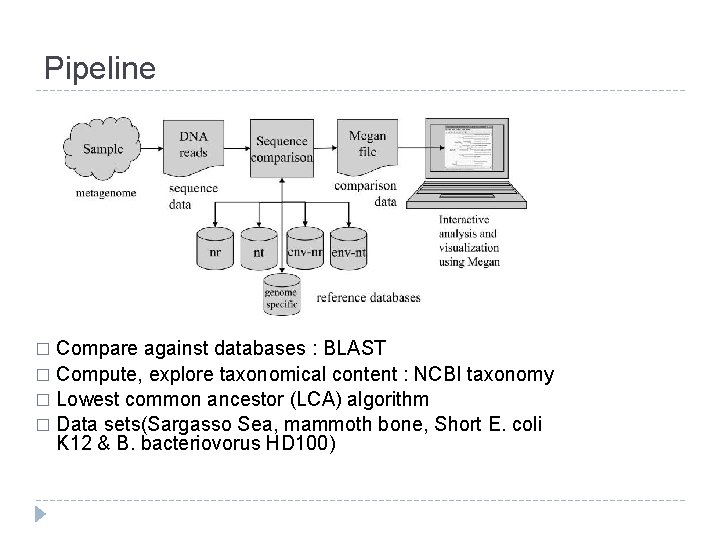
Pipeline Compare against databases : BLAST � Compute, explore taxonomical content : NCBI taxonomy � Lowest common ancestor (LCA) algorithm � Data sets(Sargasso Sea, mammoth bone, Short E. coli K 12 & B. bacteriovorus HD 100) �

What we can do with MEGAN � Species and strain identification through species-specific genes � Searching species or taxa by find tool � Distribution of strains of a species � Underlying sequence alignments

Experiments-1 � Sargasso � Sea data set � Sanger sequencing � Sample 1 -4 from DDBJ/EMBL/Gen. Bank 10000 reads from Sample 1 Randomly selected a pooled set of 10000 reads from samples 2 -4 � BLASTX->NCBI-NR � 1% no hits from sample 1, <3% no hits from sample 2 -4 � Filters � Min-score : bit-score threshold of 100 � Top-percent : bit scores lie within 5% of the best score � Min-support : isolated assignments it by one read) discarded

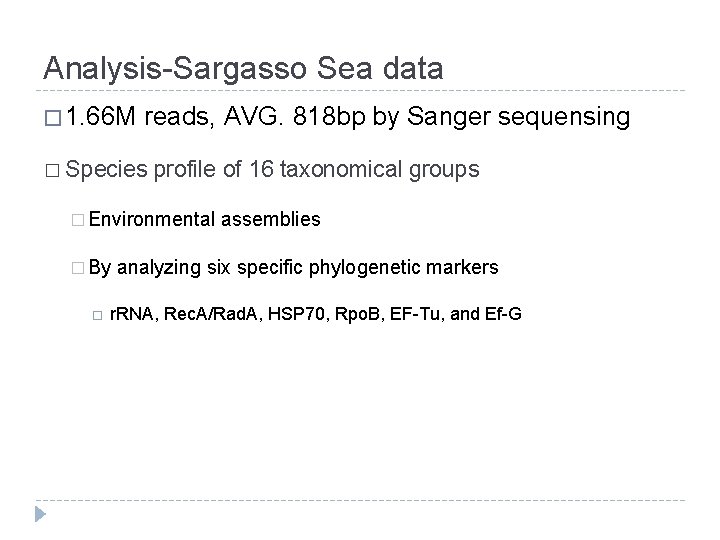
Analysis-Sargasso Sea data � 1. 66 M reads, AVG. 818 bp by Sanger sequensing � Species profile of 16 taxonomical groups � Environmental � By assemblies analyzing six specific phylogenetic markers r. RNA, Rec. A/Rad. A, HSP 70, Rpo. B, EF-Tu, and Ef-G

Result • Sample 1 • ~83% reads were assigned to taxa that were more speific than the kingdom level • Majority of (8298) were assigned to bacterial group • Sample 2 -4 • ~59% reads were assigned to taxa that were more specific than the kingdom level • Majority of (5709) were assigned to bacterial group • Alphaproteobacteria, Gammaproteobacteria by a factor of 2 -4 over the remaining 14 taxonomic groups • Eukaryotes & Viruses : size filtering • Archaea : May be there is 10 times as much vacterial sequence information in the public databases

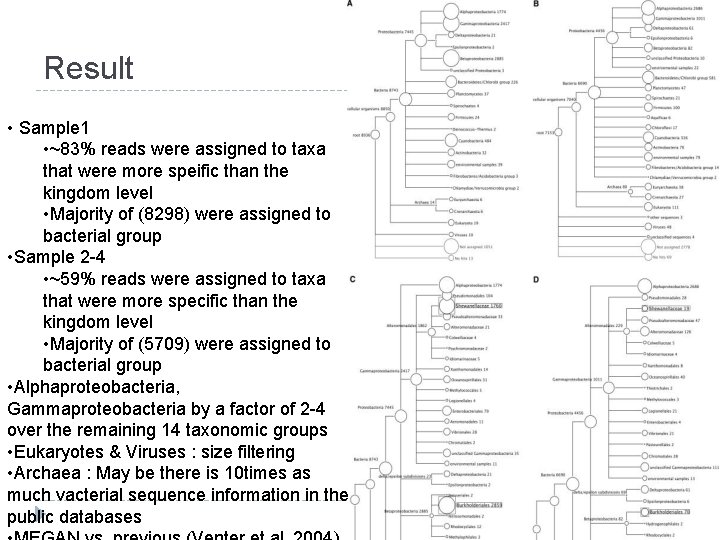
Result-cont. • Averaged weighted percentage of the siz phylogenetic markers for each of the 16 taxonomic groups • Easily detect sampling bias between sample 1 and pooled sample 2 -4

Experiments-2 � Mammoth � Data bone set � Roche GS 20 sequencing (Sequencing-by-synthesis) � Sample from 1 g of mammoth bone , 28000 years � ~300, 000 reads, 95 bp � BLASTZ-genome sequences (elephant, human, dog) � 45. 4% of the reads mammoth DNA, others are environmental organisms (bacteria, fungi, amoeba, nematodes) � BLASTX–NCBI-NR for environmental sequences � Filters : bit-score threshold 30, discard isolated assignment (filtered 2086 reads)

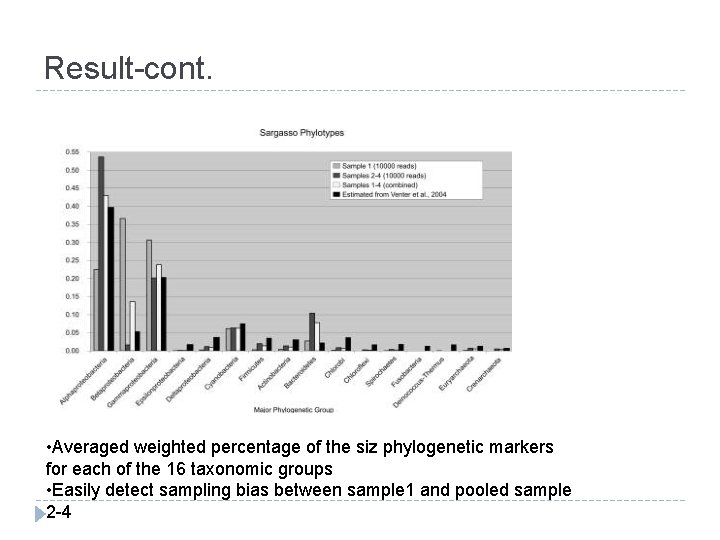
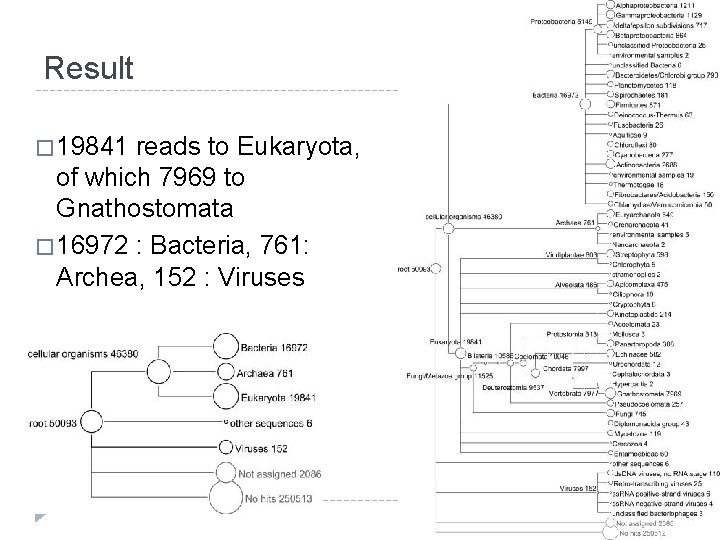
Result � 19841 reads to Eukaryota, of which 7969 to Gnathostomata � 16972 : Bacteria, 761: Archea, 152 : Viruses

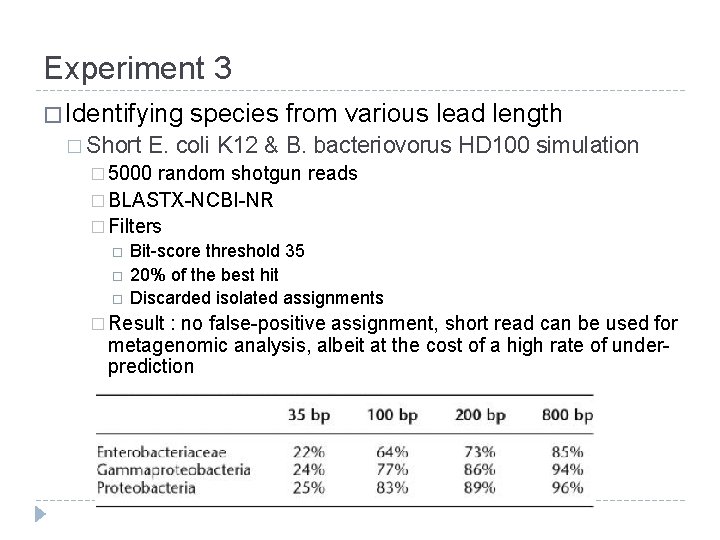
Experiment 3 � Identifying � Short species from various lead length E. coli K 12 & B. bacteriovorus HD 100 simulation � 5000 random shotgun reads � BLASTX-NCBI-NR � Filters Bit-score threshold 35 20% of the best hit Discarded isolated assignments � Result : no false-positive assignment, short read can be used for metagenomic analysis, albeit at the cost of a high rate of underprediction

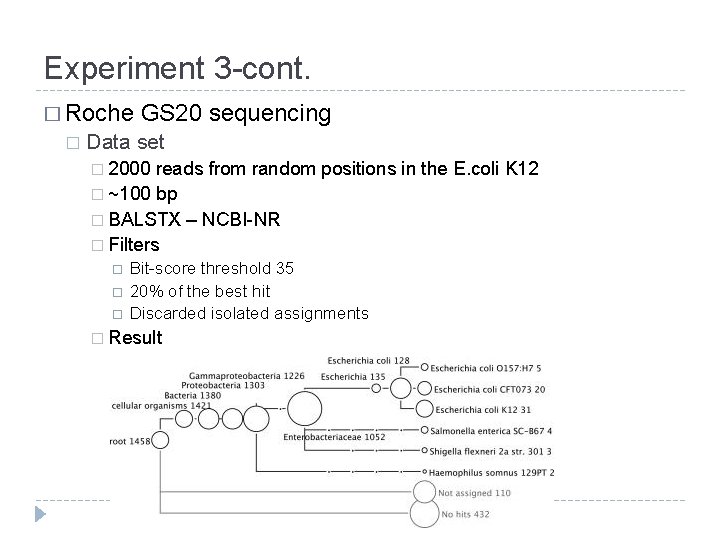
Experiment 3 -cont. � Roche � GS 20 sequencing Data set � 2000 reads from random positions in the E. coli K 12 � ~100 bp � BALSTX – NCBI-NR � Filters Bit-score threshold 35 20% of the best hit Discarded isolated assignments � Result

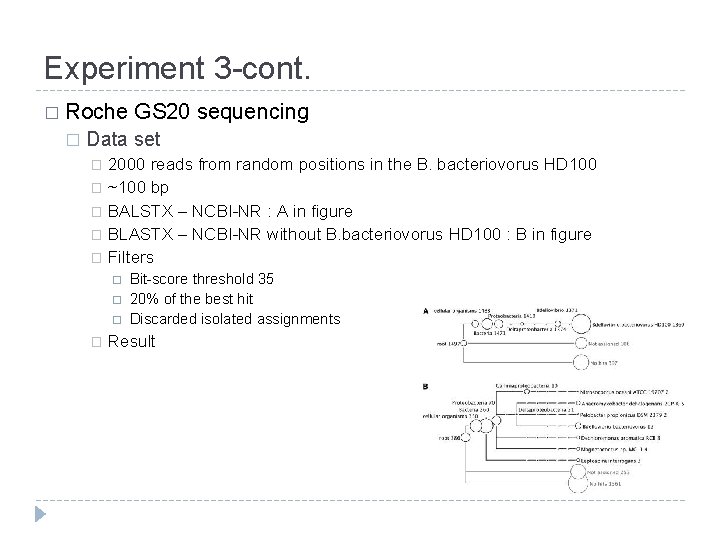
Experiment 3 -cont. � Roche � GS 20 sequencing Data set 2000 reads from random positions in the B. bacteriovorus HD 100 � ~100 bp � BALSTX – NCBI-NR : A in figure � BLASTX – NCBI-NR without B. bacteriovorus HD 100 : B in figure � Filters � � Bit-score threshold 35 20% of the best hit Discarded isolated assignments Result

MEGAN 3(June, 2009) � Suitable for very large datasets � Advances in the throughput and cost-efficiency of sequencing technology � Interests � From changed ‘which species present’ to ‘What’s different? ’ � Features � Visualization technique for multiple database � New statistical method for highlighting the difference in a pairwise comparison

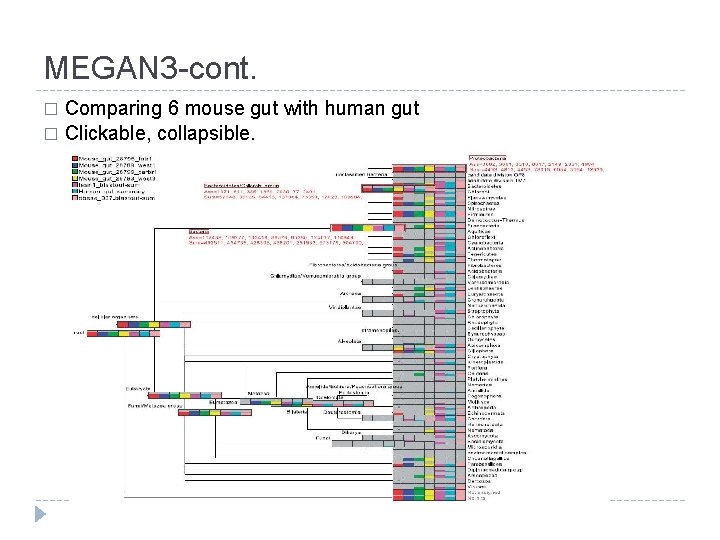
MEGAN 3 -cont. Comparing 6 mouse gut with human gut � Clickable, collapsible. �

 Curated metagenomic data
Curated metagenomic data Metagenomic binning
Metagenomic binning Dr finney unmc
Dr finney unmc Megan and ron ate too much
Megan and ron ate too much Megan meiers
Megan meiers Megan kosirog md
Megan kosirog md Fortran c interoperability
Fortran c interoperability Cyber career pathways tool
Cyber career pathways tool Naughtia childs porno
Naughtia childs porno Megan andring
Megan andring Jesse timmendequas
Jesse timmendequas Megan madrigal md
Megan madrigal md Megan templin
Megan templin Anabel rojas
Anabel rojas Megan suelzer
Megan suelzer Megan koeth
Megan koeth Megan baines
Megan baines Megan e
Megan e