A Prionlike Mechanism in Amyotrophic Lateral Sclerosis Dr


- Slides: 12

A Prion-like Mechanism in Amyotrophic Lateral Sclerosis Dr. Eamonn F. Healy Professor of Chemistry St. Edward’s University Austin, Tx.

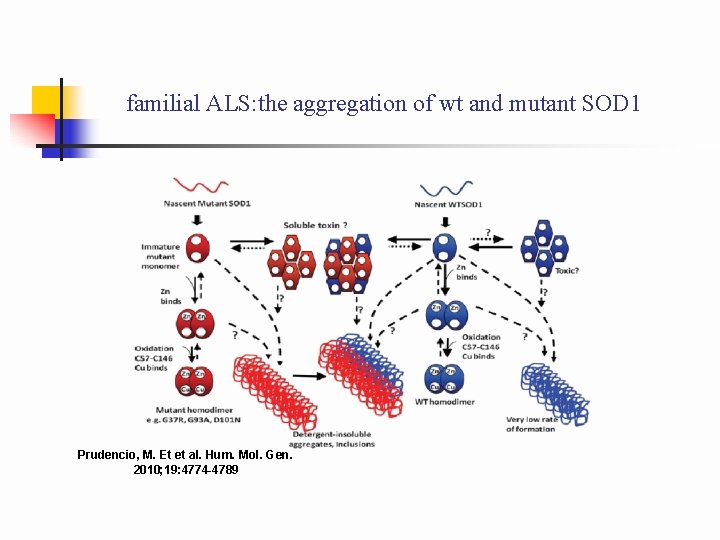
familial ALS: the aggregation of wt and mutant SOD 1 Prudencio, M. Et et al. Hum. Mol. Gen. 2010; 19: 4774 -4789

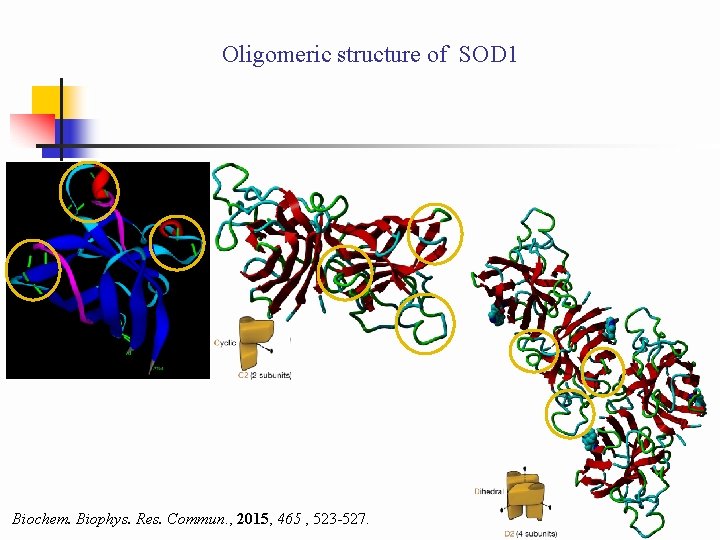
Oligomeric structure of SOD 1 Biochem. Biophys. Res. Commun. , 2015, 465 , 523 -527.

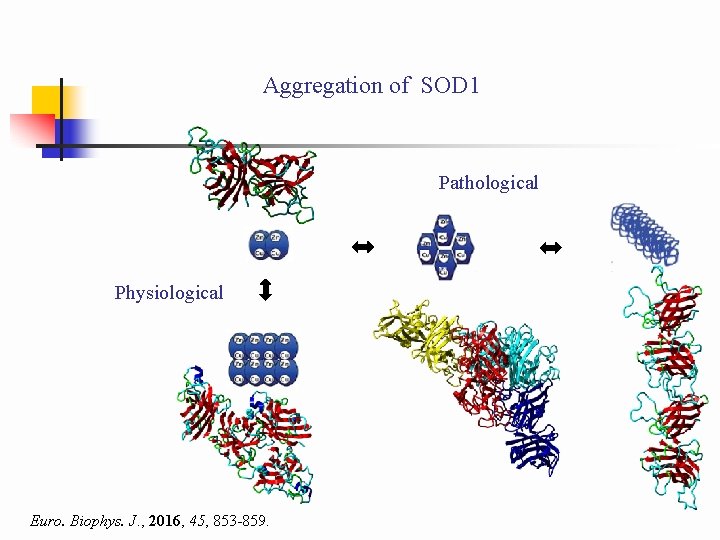
Aggregation of SOD 1 Pathological Physiological Euro. Biophys. J. , 2016, 45, 853 -859.

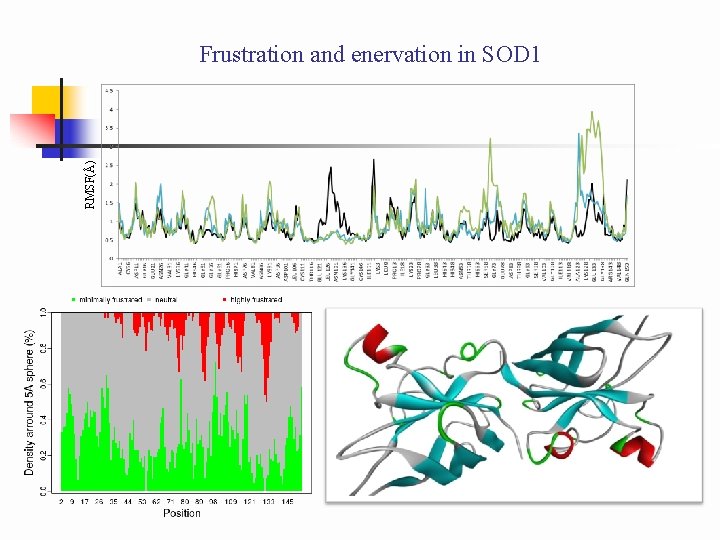
RMSF(Å) Frustration and enervation in SOD 1

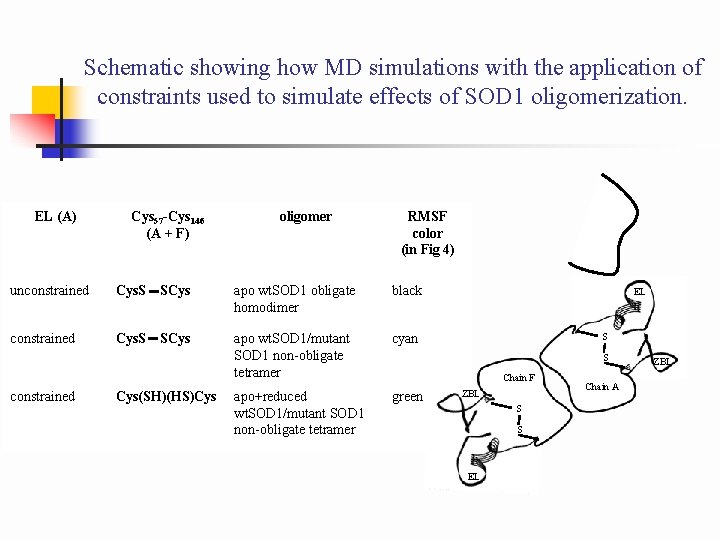
Schematic showing how MD simulations with the application of constraints used to simulate effects of SOD 1 oligomerization. EL (A) Cys 57 -Cys 146 (A + F) oligomer RMSF color (in Fig 4) unconstrained Cys. S SCys apo wt. SOD 1 obligate homodimer black constrained Cys. S SCys apo wt. SOD 1/mutant SOD 1 non-obligate tetramer cyan apo+reduced wt. SOD 1/mutant SOD 1 non-obligate tetramer green constrained Cys(SH)(HS)Cys EL S S Chain F ZBL S S EL Chain A ZBL

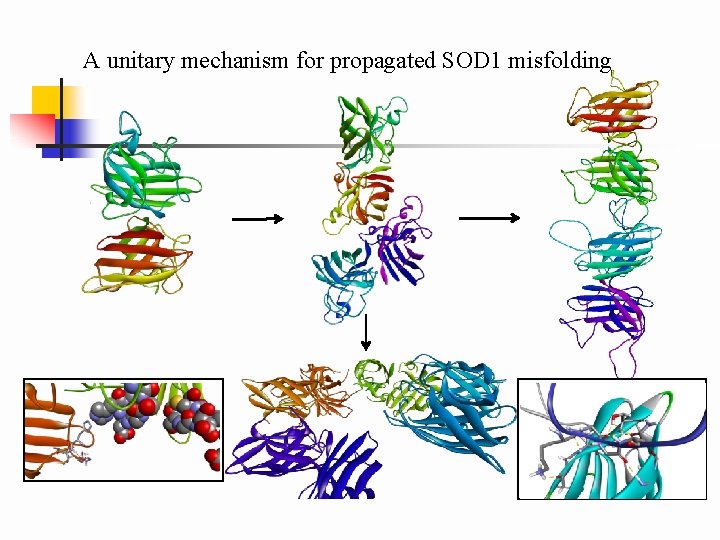
A unitary mechanism for propagated SOD 1 misfolding

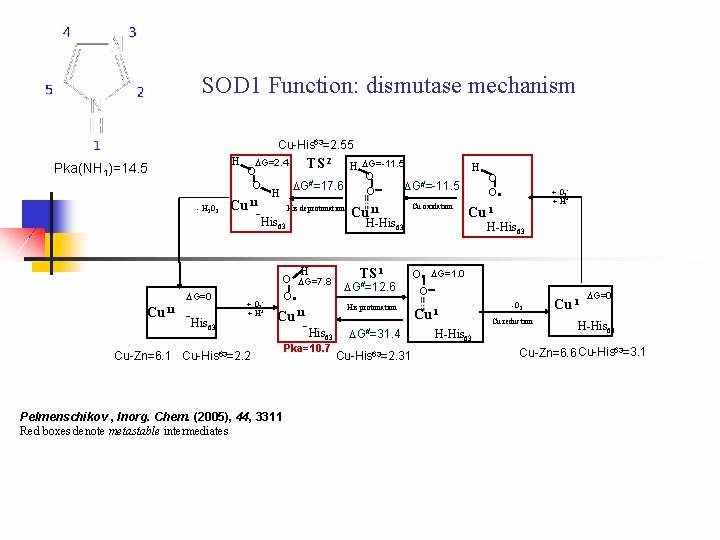
SOD 1 Function: dismutase mechanism Cu-His 63=2. 55 H Pka(NH 1)=14. 5 - H 2 O 2 DG=2. 4 O O Cu II H DG=-11. 5 O DG#=17. 6 DG#=-11. 5 O - H -His TS 2 His deprotonation Cu II - His 63 + O 2 -. + H+ H O DG=7. 8 O . Cu II -His 63 Cu-Zn=6. 1 Cu-His 63=2. 2 Pelmenschikov , Inorg. Chem. (2005), 44, 3311 Red boxes denote metastable intermediates Cu oxidation H-His 63 63 DG=0 Cu II Pka=10. 7 TS 1 DG#=12. 6 His protonation DG#=31. 4 Cu-His 63=2. 31 H O O . + O 2 -. + H+ Cu I H-His 63 . O DG=1. 0 O - Cu I H-His 63 -O 2 Cu reduction Cu I DG=0 H-His 63 Cu-Zn=6. 6 Cu-His 63=3. 1

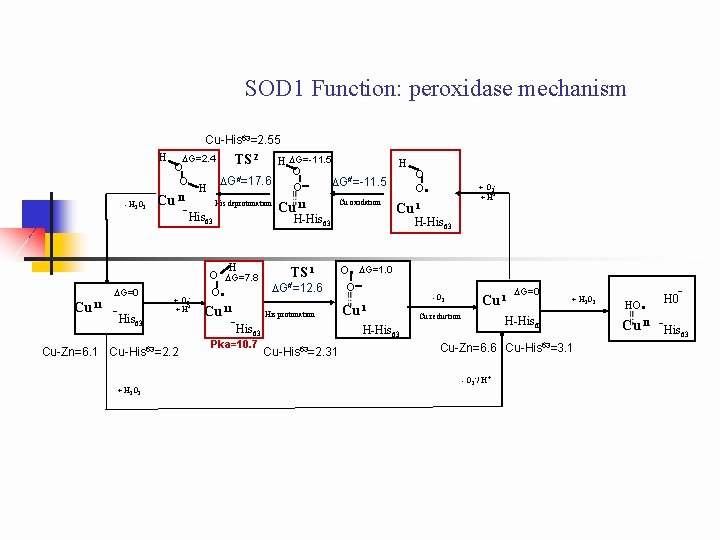
SOD 1 Function: peroxidase mechanism Cu-His 63=2. 55 H - H 2 O 2 DG=2. 4 O O Cu II H DG=-11. 5 O DG#=17. 6 DG#=-11. 5 O His deprotonation Cu II - His 63 + O 2 -. + H+ Cu II H O DG=7. 8 O . Cu II -His TS 1 DG#=12. 6 His protonation + H 2 O 2 Pka=10. 7 O O . + O 2 -. + H+ Cu I H-His 63 . O DG=1. 0 O - Cu I -O 2 Cu reduction Cu-His 63=2. 31 DG=0 + H 2 O 2 H-His 63 63 Cu-Zn=6. 1 Cu-His 63=2. 2 Cu oxidation H-His 63 63 DG=0 H -His TS 2 Cu-Zn=6. 6 Cu-His 63=3. 1 - O 2 -/ H+. HO . - H 0 Cu II -His 63

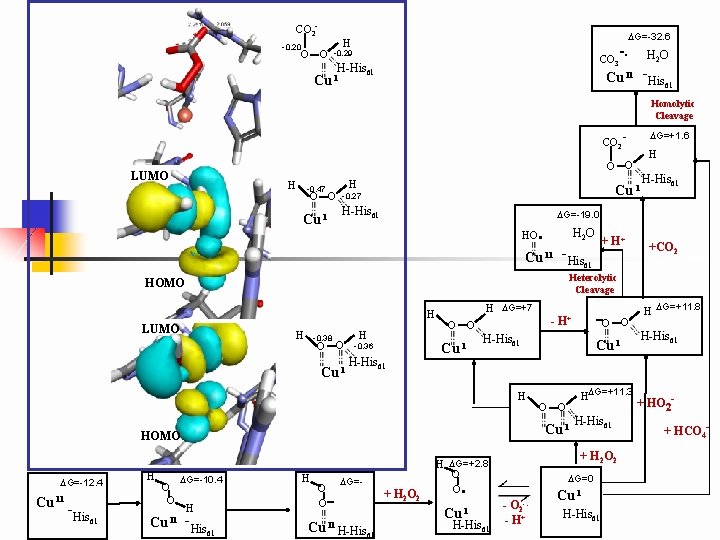
CO 2 - DG=-32. 6 H -0. 20 -. CO O O -0. 29 H 2 O 3 H-His 61 Cu II -His 61 Cu I Homolytic Cleavage - DG=+1. 6 CO 2 LUMO H H -0. 47 O O -0. 27 Cu I H-His 61 HO . H 2 O + H+ H LUMO H H -0. 38 O O -0. 36 Cu I H-His 61 H DG=+7 O O Cu I -His 61 Cu II H -His 61 H O O - DG=- Cu II H-His 61 + H 2 O 2 O O . H-His 61 + H 2 O 2 H DG=+2. 8 O O Cu I O HDG=+11. 3 Cu I DG=-10. 4 -O Cu I HOMO Cu II - H+ H-His 61 H O O +CO 2 Heterolytic Cleavage HOMO H H-His 61 DG=-19. 0 Cu II -His 61 DG=-12. 4 H O O DG=0 - O 2 - H+ -. Cu I H-His 61 H DG=+11. 8 H-His 61 + HO 2+ HCO 4 -

End on binding of hydrogen peroxide His 61 His 46 His 118 His 44

Regulation of activity through competitive inhibition
 Ms title
Ms title Scleroderma
Scleroderma Multiple sclerosis concept map
Multiple sclerosis concept map Hodgkin lymphoma nodular sclerosis
Hodgkin lymphoma nodular sclerosis Cell
Cell Meggyógyultam a sclerosis multiplexből
Meggyógyultam a sclerosis multiplexből Concept map of assessment
Concept map of assessment How to find total surface area of prism
How to find total surface area of prism Jalur karir keterampilan lateral
Jalur karir keterampilan lateral Tronco parte del cuerpo
Tronco parte del cuerpo Pensamiento lateral
Pensamiento lateral Ridge parallelism
Ridge parallelism Lateral faces of cone
Lateral faces of cone