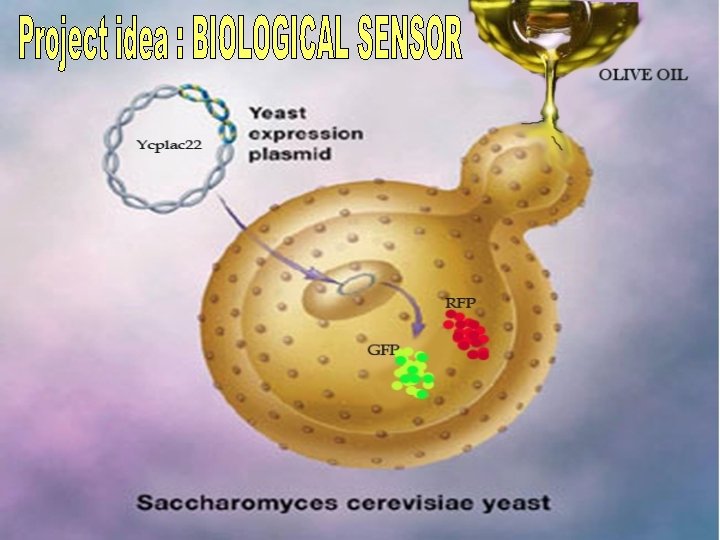
Yes Oil ENGINEERED YEAST CELLS A Yeast Sensor



![BIOLOGICAL CIRCUIT [O. A]<2. 8 m. M [O. A. ]>7. 1 m. M RFP BIOLOGICAL CIRCUIT [O. A]<2. 8 m. M [O. A. ]>7. 1 m. M RFP](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-10.jpg)
![WHAT HAPPENS? Virgin Oil 2. 8 m. M<[O. A]<7. 1 m. M WHAT HAPPENS? Virgin Oil 2. 8 m. M<[O. A]<7. 1 m. M](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-11.jpg)
![OUTPUTS [Oleic acid] < 2, 8 m. M Extra Virgin Olive Oil [Oleic acid] OUTPUTS [Oleic acid] < 2, 8 m. M Extra Virgin Olive Oil [Oleic acid]](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-13.jpg)

![Mathematical Model Inputs d Degradations Pho 4 p dt SA αA [PHO 4 p] Mathematical Model Inputs d Degradations Pho 4 p dt SA αA [PHO 4 p]](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-21.jpg)

- Slides: 31

Yes. Oil ENGINEERED YEAST CELLS: A Yeast Sensor for real Extra Virgin Olive Oil Federico II University

Irene, biologist Roberta, biologist Giovanni, engineer Lucia, mathematician Giulia, biologist Velia, biologist Maria Aurelia, biologist Alda, biologist

We are from… ITAL Y NAPLES


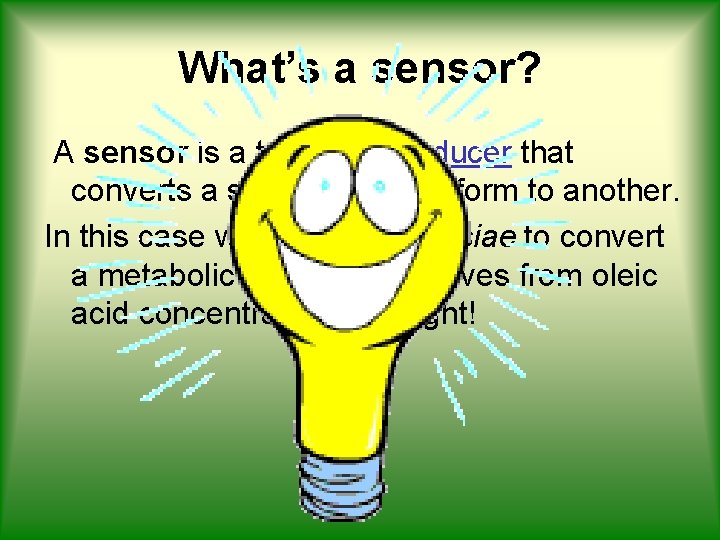
What’s a sensor? A sensor is a type of transducer that converts a signal from one form to another. In this case we want S. cerevisiae to convert a metabolic signal, that derives from oleic acid concentration, into light!

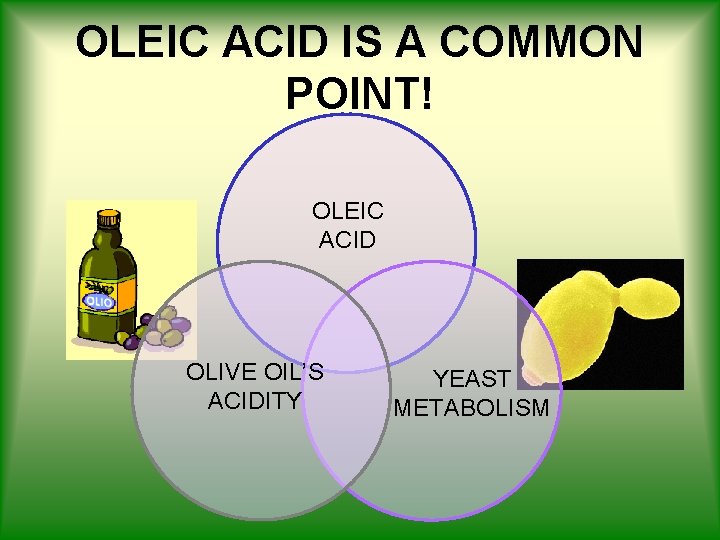
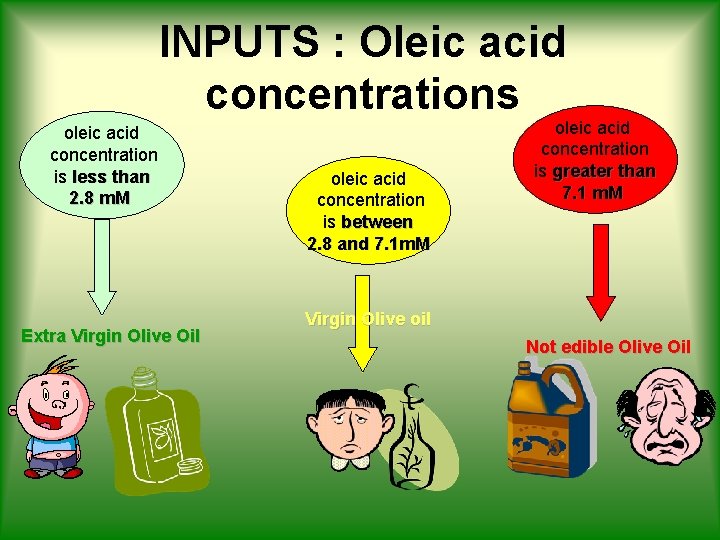
Background • Oleic acid is the principal component of Olive Oil • It’s the indicator of Oil’s acidity Olive Oil is classified as follows: ü [oleic acid] < 2. 8 m. M EXTRA VIRGIN ü [oleic acid] < 7. 1 m. M VIRGIN ü [oleic acid] > 7. 1 m. M NOT EDIBLE

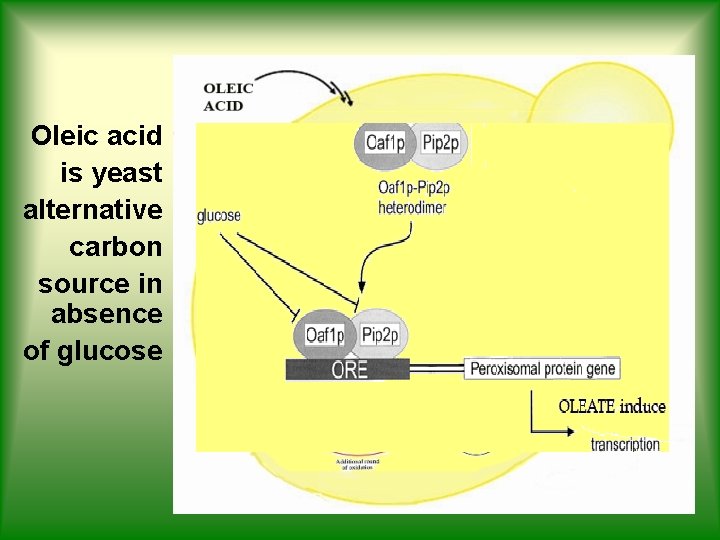
Oleic acid is yeast alternative carbon source in absence of glucose

OLEIC ACID IS A COMMON POINT! OLEIC ACID OLIVE OIL’S ACIDITY YEAST METABOLISM

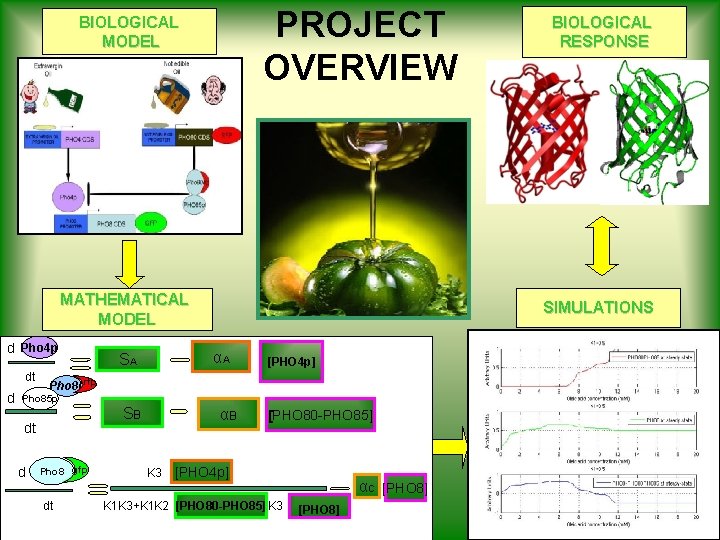
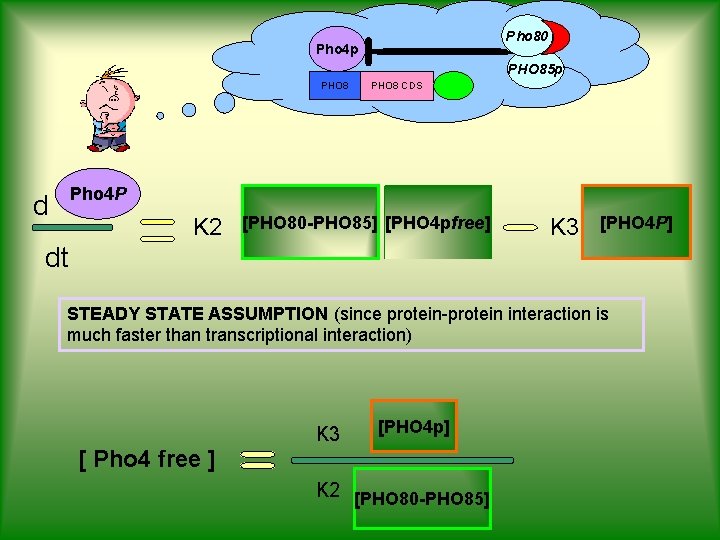
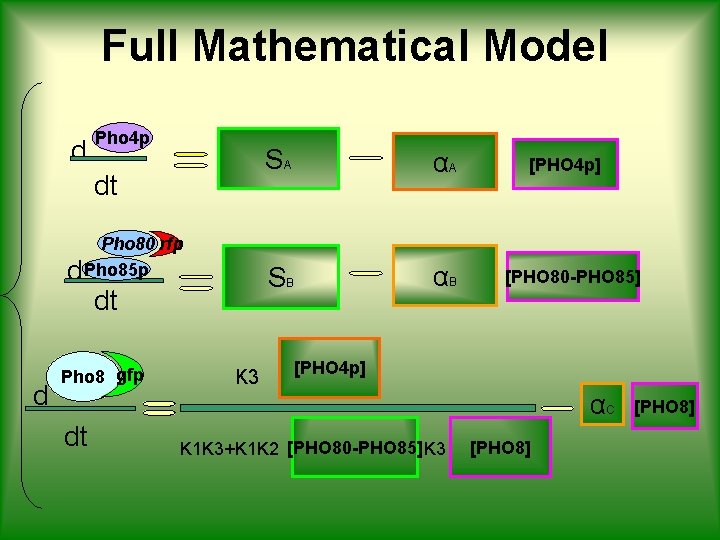
PROJECT OVERVIEW BIOLOGICAL MODEL MATHEMATICAL MODEL d Pho 4 p dt d Pho 80 rfp Pho 85 p dt d Pho 8 gfp dt SIMULATIONS αA SA SB αB K 3 BIOLOGICAL RESPONSE [PHO 4 p] [PHO 80 -PHO 85] [PHO 4 p] K 1 K 3+K 1 K 2 [PHO 80 -PHO 85] K 3 αC [PHO 8]
![BIOLOGICAL CIRCUIT O A2 8 m M O A 7 1 m M RFP BIOLOGICAL CIRCUIT [O. A]<2. 8 m. M [O. A. ]>7. 1 m. M RFP](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-10.jpg)
BIOLOGICAL CIRCUIT [O. A]<2. 8 m. M [O. A. ]>7. 1 m. M RFP
![WHAT HAPPENS Virgin Oil 2 8 m MO A7 1 m M WHAT HAPPENS? Virgin Oil 2. 8 m. M<[O. A]<7. 1 m. M](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-11.jpg)
WHAT HAPPENS? Virgin Oil 2. 8 m. M<[O. A]<7. 1 m. M

INPUTS : Oleic acid concentrations oleic acid concentration is less than 2. 8 m. M Extra Virgin Olive Oil oleic acid concentration is between 2. 8 and 7. 1 m. M oleic acid concentration is greater than 7. 1 m. M Virgin Olive oil Not edible Olive Oil
![OUTPUTS Oleic acid 2 8 m M Extra Virgin Olive Oil Oleic acid OUTPUTS [Oleic acid] < 2, 8 m. M Extra Virgin Olive Oil [Oleic acid]](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-13.jpg)
OUTPUTS [Oleic acid] < 2, 8 m. M Extra Virgin Olive Oil [Oleic acid] > 7, 1 m. M Virgin Oil Not edible Olive Oil

Biological Model v. What microorganism? v. Which promoters? v. Which genes?

Choice of microorganism: S. cerevisiae 1. In yeast genome there are genes activated by Oleic Acid. 2. Yeast is not a dangerous microorganism for consumers that will use the oil 3. Yeast can be easily engineered

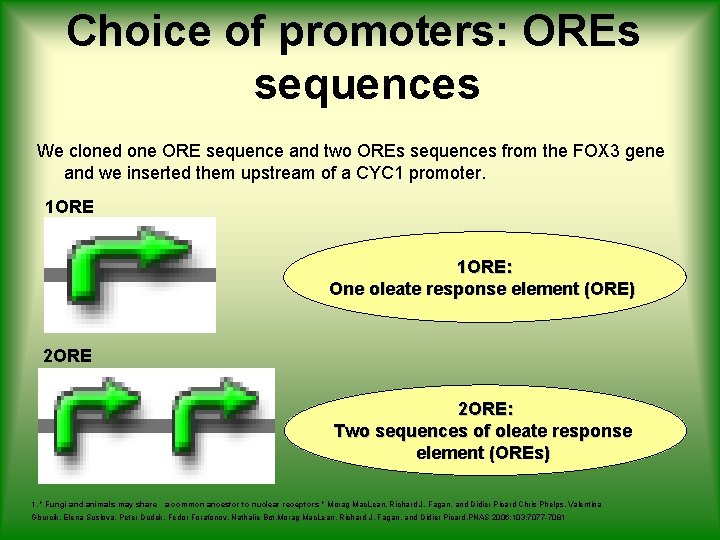
Choice of promoters: OREs sequences We cloned one ORE sequence and two OREs sequences from the FOX 3 gene and we inserted them upstream of a CYC 1 promoter. 1 ORE: One oleate response element (ORE) 2 ORE: Two sequences of oleate response element (OREs) 1. " Fungi and animals may share a common ancestor to nuclear receptors " Morag Mac. Lean, Richard J. Fagan, and Didier Picard Chris Phelps, Valentina Gburcik, Elena Suslova, Peter Dudek, Fedor Forafonov, Nathalie Bot, Morag Mac. Lean, Richard J. Fagan, and Didier Picard. PNAS 2006; 103; 7077 -7081

Choice of genes : PHO-patway PHO 8 p-GFP PHO 4 p PHO 8 PROMOTER PHO 80 p-RFP PHO 85 p PHO 4 p

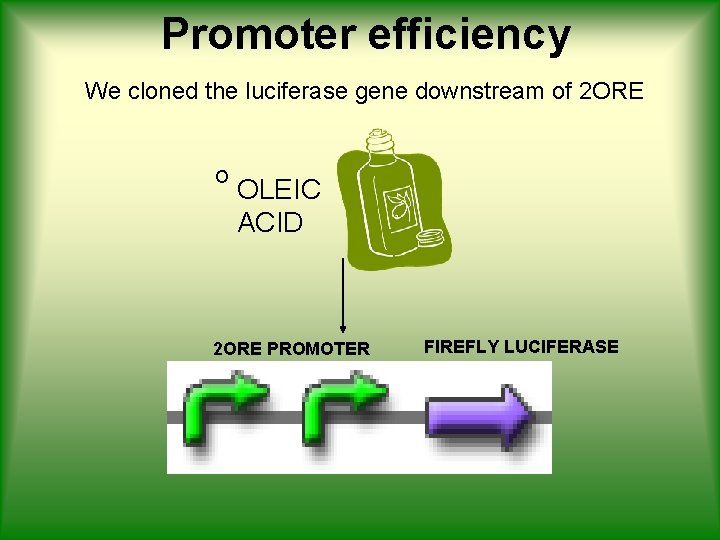
Promoter efficiency We cloned the luciferase gene downstream of 2 ORE O OLEIC ACID 2 ORE PROMOTER FIREFLY LUCIFERASE

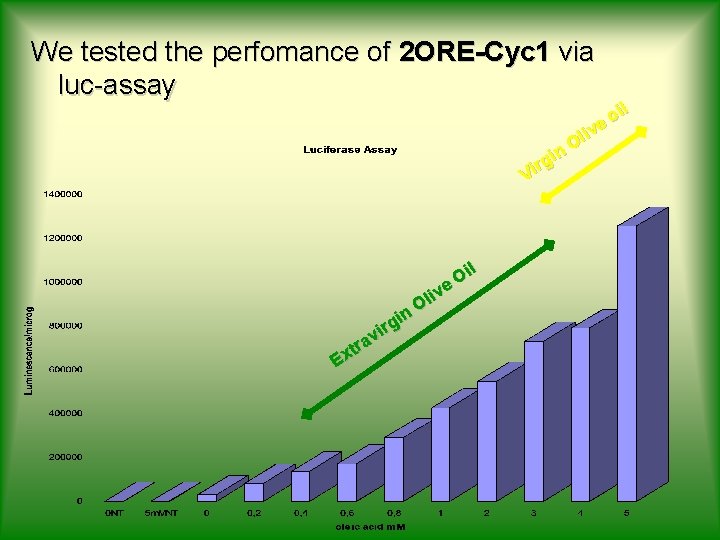
We tested the perfomance of 2 ORE-Cyc 1 via luc-assay il o ive l O n i rg Vi il O ive l O n i irg v tra x E

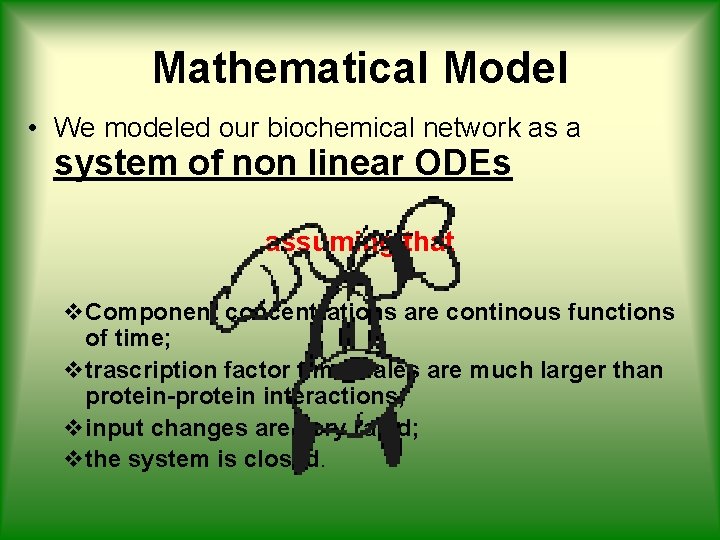
Mathematical Model • We modeled our biochemical network as a system of non linear ODEs assuming that v. Component concentrations are continous functions of time; vtrascription factor timescales are much larger than protein-protein interactions; vinput changes are very rapid; vthe system is closed.
![Mathematical Model Inputs d Degradations Pho 4 p dt SA αA PHO 4 p Mathematical Model Inputs d Degradations Pho 4 p dt SA αA [PHO 4 p]](https://slidetodoc.com/presentation_image/b580982920cc23cdb19e0f7c049c9387/image-21.jpg)
Mathematical Model Inputs d Degradations Pho 4 p dt SA αA [PHO 4 p] SB αB [PHO 80 -PHO 85] Pho 80 rfp d PHO 85 p dt d PHO 8 gfp dt [ Pho 4 free ] K 1 + [Pho 4 free] Michaelis-Menten Term αC [PHO 8]

Pho 80 Pho 4 p PHO 85 p PHO 8 CDS Pho 4 P d K 2 [PHO 80 -PHO 85] [PHO 4 pfree] K 3 [PHO 4 P] dt STEADY STATE ASSUMPTION (since protein-protein interaction is much faster than transcriptional interaction) K 3 [PHO 4 p] [ Pho 4 free ] K 2 [PHO 80 -PHO 85]

Full Mathematical Model d Pho 4 p dt Pho 80 rfp d Pho 85 p dt d Pho 8 gfp K 3 SA αA SB αB [PHO 4 p] [PHO 80 -PHO 85] [PHO 4 p] αC dt K 1 K 3+K 1 K 2 [PHO 80 -PHO 85] K 3 [PHO 8]

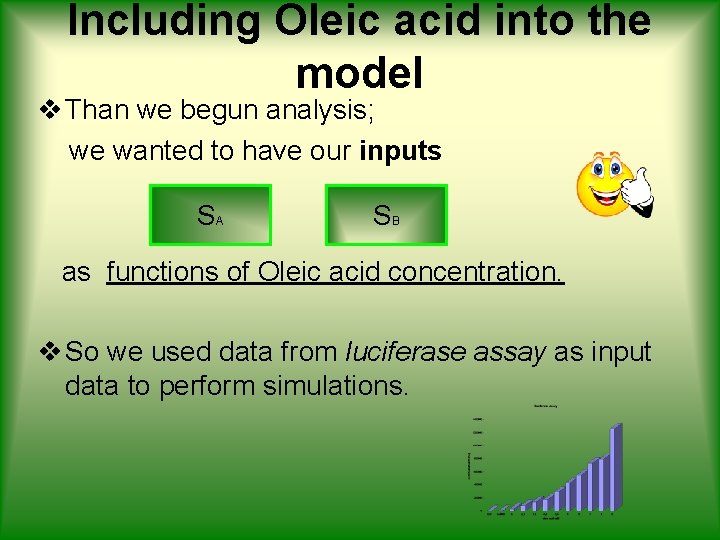
Including Oleic acid into the model v Than we begun analysis; we wanted to have our inputs SA SB as functions of Oleic acid concentration. v So we used data from luciferase assay as input data to perform simulations.

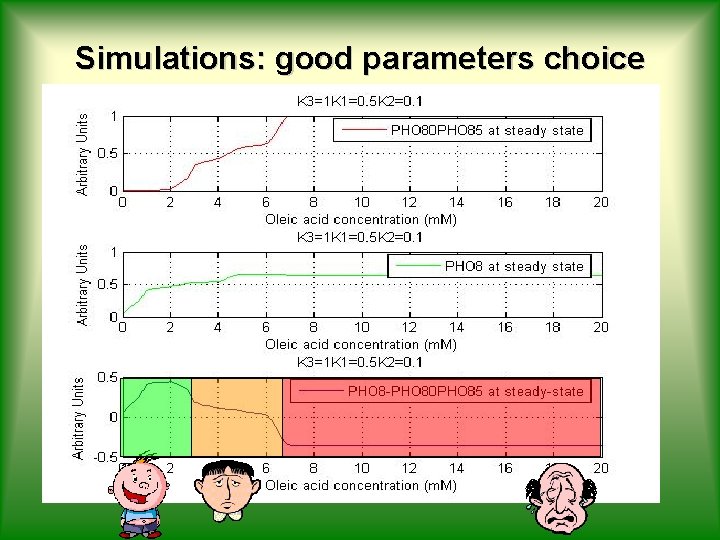
Simulations: good parameters choice

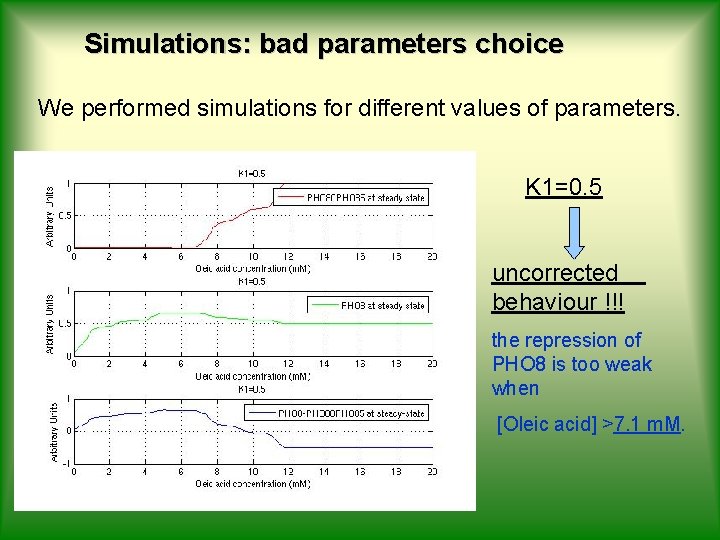
Simulations: bad parameters choice We performed simulations for different values of parameters. K 1=0. 5 uncorrected behaviour !!! the repression of PHO 8 is too weak when [Oleic acid] >7. 1 m. M.

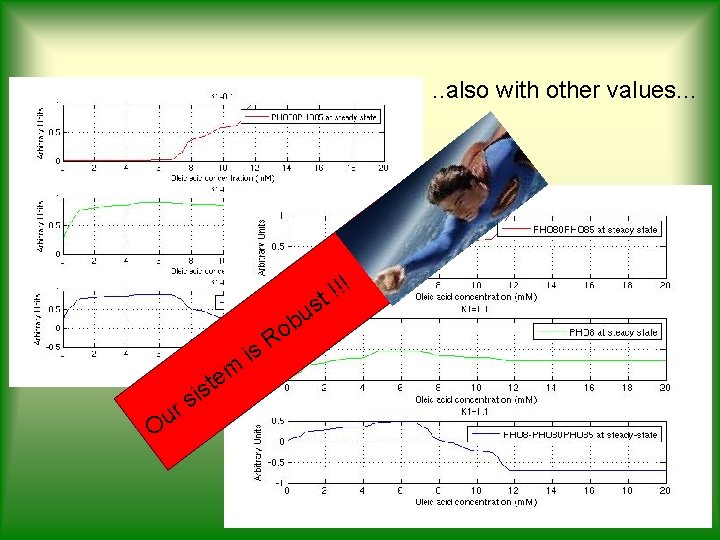
. . also with other values. . . ! o R is m e r u O t s i s !! t s u b

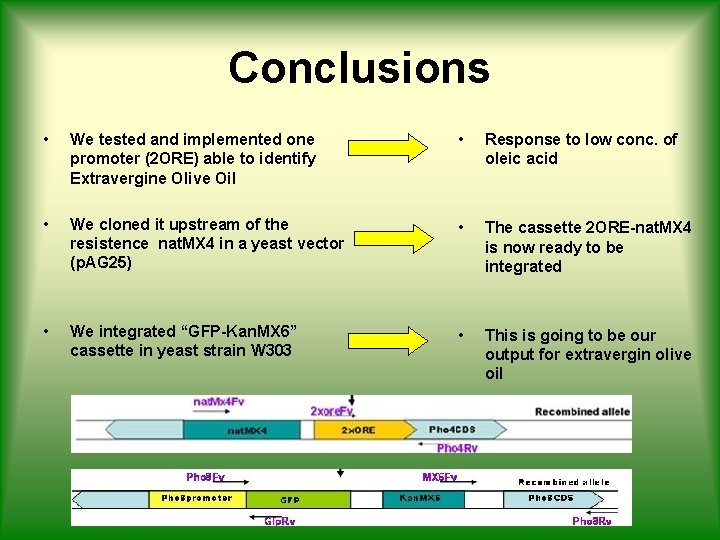
Conclusions • We tested and implemented one promoter (2 ORE) able to identify Extravergine Olive Oil • Response to low conc. of oleic acid • We cloned it upstream of the resistence nat. MX 4 in a yeast vector (p. AG 25) • The cassette 2 ORE-nat. MX 4 is now ready to be integrated • We integrated “GFP-Kan. MX 6” cassette in yeast strain W 303 • This is going to be our output for extravergin olive oil

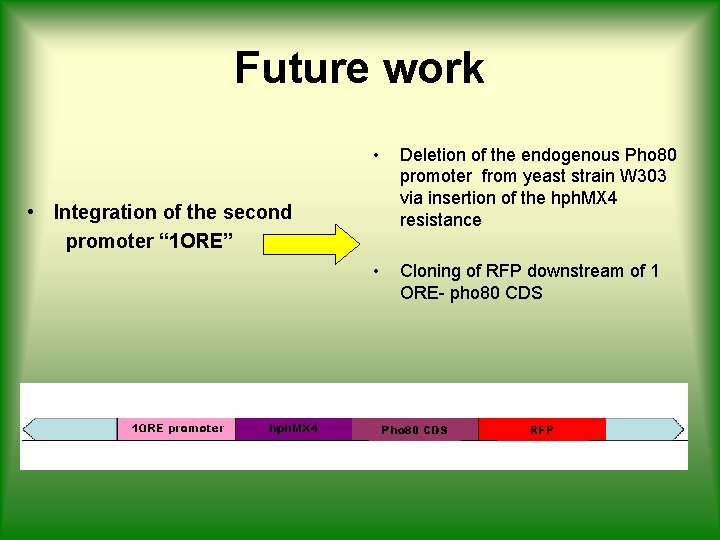
Future work • Deletion of the endogenous Pho 80 promoter from yeast strain W 303 via insertion of the hph. MX 4 resistance • Cloning of RFP downstream of 1 ORE- pho 80 CDS • Integration of the second promoter “ 1 ORE”

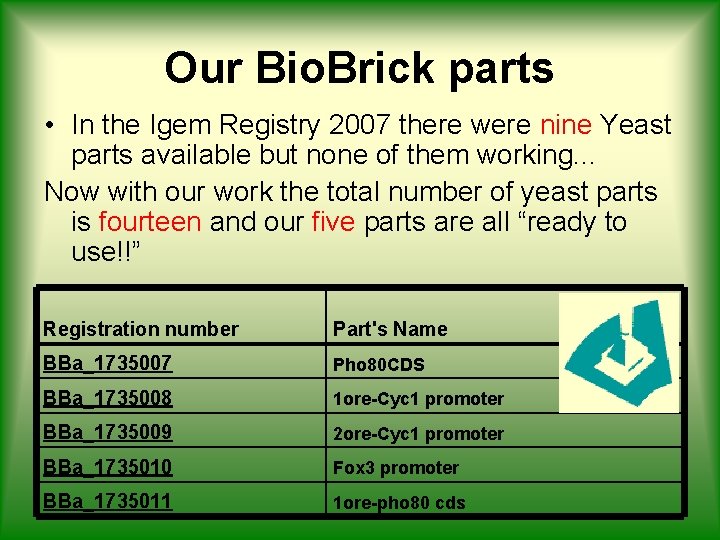
Our Bio. Brick parts • In the Igem Registry 2007 there were nine Yeast parts available but none of them working. . . Now with our work the total number of yeast parts is fourteen and our five parts are all “ready to use!!” Registration number Part's Name BBa_1735007 Pho 80 CDS BBa_1735008 1 ore-Cyc 1 promoter BBa_1735009 2 ore-Cyc 1 promoter BBa_1735010 Fox 3 promoter BBa_1735011 1 ore-pho 80 cds

Thanks to. . . Instructors: Maria Pia Cosma Diego di Bernardo Mario di Bernardo