YEAST MOLECULAR GENETICS A Yeast genetics nomenclature w









- Slides: 34

YEAST MOLECULAR GENETICS (A)

Yeast genetics: nomenclature w w w w Yeast genes have names consisting of three letters and up to three numbers: GPD 1, HSP 12, PDC 6. . . Usually they are meaningful (or meaningless) abbreviations Wild type genes are written with capital letters in italics: TPS 1, RHO 1, CDC 28. . . Recessive mutant genes are written with small letters in italics: tps 1, rho 1, cdc 28 Mutant alleles are designated with a dash and a number: tps 1 -1, rho 1 -23, cdc 28 -2 If the mutation has been constructed, i. e. by gene deletion, this is indicated and the genetic marker used for deletion too: tps 1 D: : HIS 3 The gene product, a protein, is written with a capital letter at the beginning and not in italics; often a ”p” is added at the end: Tps 1 p, Rho 1 p, Cdc 28 p Many genes have of course only be found by systematic sequencing and as long as their function is not determined they get a landmark name: YDR 518 C, YML 016 W. . . , where n n © Stefan Hohmann 2000 -2004 n n n w Y stands for ”yeast” The second letter represents the chromosome (D=IV, M=XIII. . ) L or R stand for left or right chromosome arm The three-digit number stands for the ORF counted from the centromere on that chromosome arm C or W stand for ”Crick” or ”Watson”, i. e. indicate the strand or direction of the ORF Some genes do not follow this nomenclature: you heard already about: HO, MATa

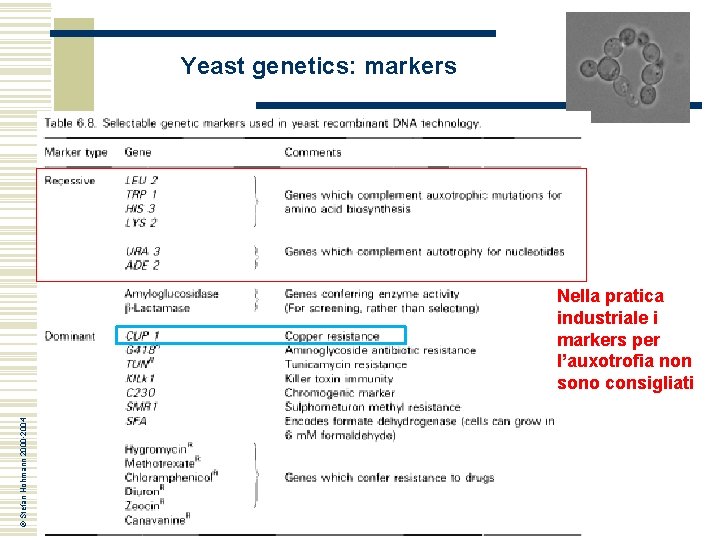
Yeast genetics: markers and strains w w Genetic markers are used to follow chromosomes in genetic crosses, to select diploids in genetic crosses, to select transformants in transformation with plasmids or integration of genes into the genome Commonly genetic markers cause auxotrophies: HIS 3, URA 3, TRP 1, LEU 2, LYS 2, ADE 2 The ade 2 mutation has a specific useful feature: cells turn red The first markers in yeast genetics were fermentation markers, i. e. genes that confer the ability to catabolise certain substrates: SUC, MAL, GAL © Stefan Hohmann 2000 -2004 GENETIC MARKERS AND STRAINS ARE TWO ELEMENTS OF THE SAME SYSTEM How can be selected a mutant yeast strain and the complementing wild type gene? ? ?

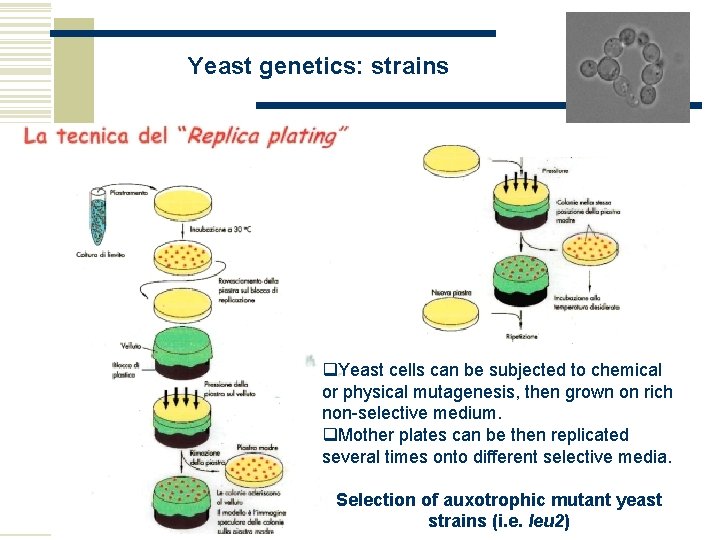
© Stefan Hohmann 2000 -2004 Yeast genetics: strains q. Yeast cells can be subjected to chemical or physical mutagenesis, then grown on rich non-selective medium. q. Mother plates can be then replicated several times onto different selective media. Selection of auxotrophic mutant yeast strains (i. e. leu 2)

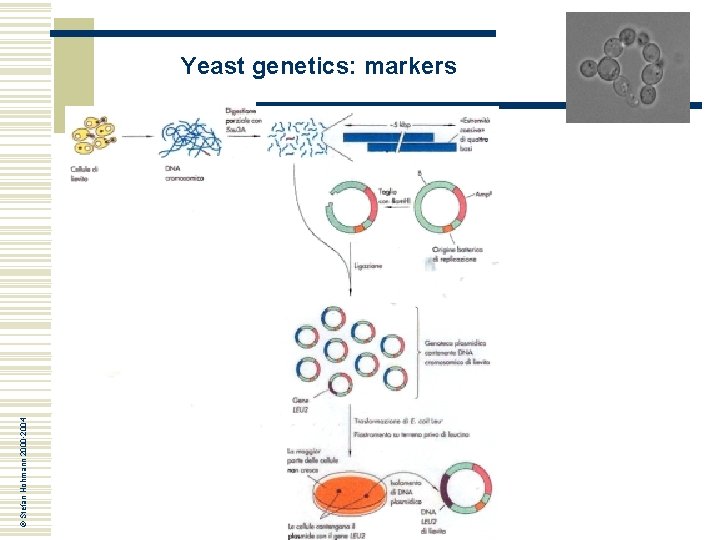
Yeast genetics: markers w Once selected yeast leu 2 mutant cells, how can we select the wild type “complementing“ gene? ? ? © Stefan Hohmann 2000 -2004 Functional complementation in E. coli leu 2 mutant cells!!!

© Stefan Hohmann 2000 -2004 Yeast genetics: markers

© Stefan Hohmann 2000 -2004

© Stefan Hohmann 2000 -2004

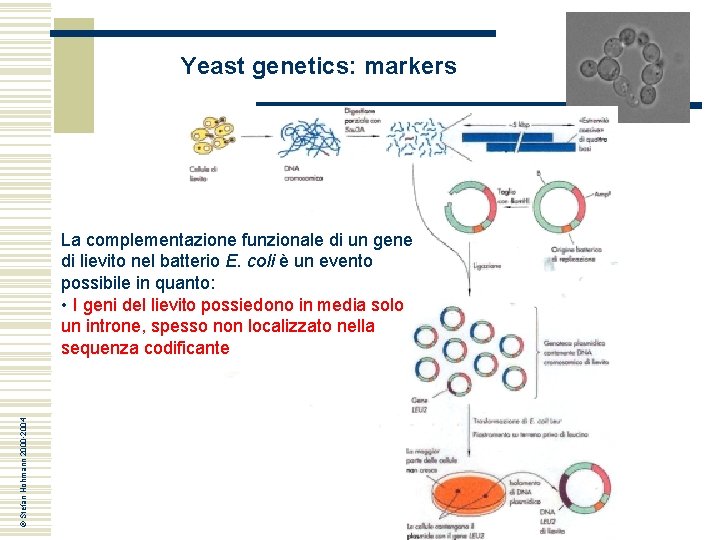
Yeast genetics: markers © Stefan Hohmann 2000 -2004 La complementazione funzionale di un gene di lievito nel batterio E. coli è un evento possibile in quanto: • I geni del lievito possiedono in media solo un introne, spesso non localizzato nella sequenza codificante

Yeast genetics: markers • I geni del lievito possiedono in media solo un introne, spesso non localizzato nella sequenza codificante © Stefan Hohmann 2000 -2004 However, in yeast only 283 of the 6000 genes contain introns and their impact on cell function is not clear. To assess the contribution of introns to cell function, large-scale intron deletions in yeast was carried out with the ultimate goal of creating an intron-free model eukaryote. It was shown that about one-third of yeast introns are not essential for growth. Only three intron deletions caused severe growth defects, but normal growth was restored in all cases by expressing the intronless m. RNA from a heterologous promoter. Twenty percent of the intron deletions caused minor phenotypes under different growth conditions. Strikingly, the combined deletion of all introns from the 15 cytoskeleton-related genes did not affect growth or strain fitness. Therefore, although the presence of introns may optimize gene expression and provide benefit under stress, a majority of introns could be removed with minor consequences on growth under laboratory conditions, supporting the view that many introns could be phased out of Saccharomyces cerevisiae without blocking cell growth. Parenteau J et al. , 2008 Mol Biol Cell. 2008 May; 19(5): 1932– 1941

© Stefan Hohmann 2000 -2004 Yeast genetics: markers La complementazione funzionale di un gene di lievito nel batterio E. coli è un evento possibile in quanto: • I geni del lievito possiedono in media solo un introne, spesso non localizzato nella sequenza codificante • Intorno all’ATG dei geni eucariotici ci sono sequenze analoghe alla Shine-Dalgarno procariotica (traducibilità) Kozak consensus sequence

© Stefan Hohmann 2000 -2004 Yeast genetics: markers

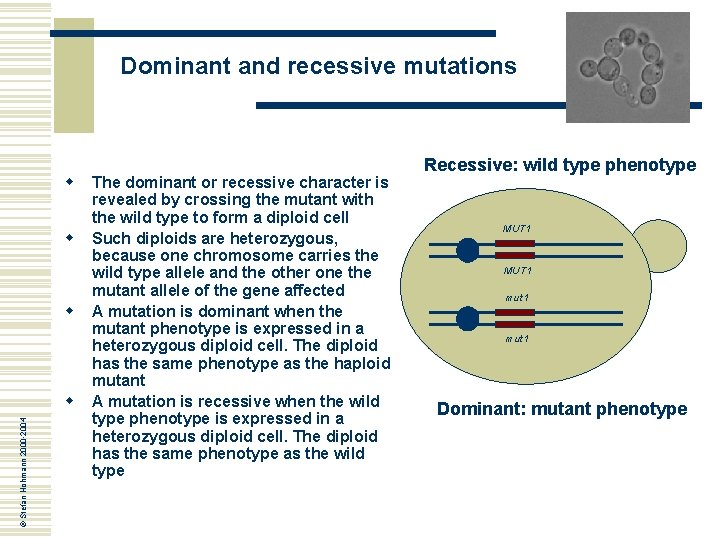
Dominant and recessive mutations w w w © Stefan Hohmann 2000 -2004 w The dominant or recessive character is revealed by crossing the mutant with the wild type to form a diploid cell Such diploids are heterozygous, because one chromosome carries the wild type allele and the other one the mutant allele of the gene affected A mutation is dominant when the mutant phenotype is expressed in a heterozygous diploid cell. The diploid has the same phenotype as the haploid mutant A mutation is recessive when the wild type phenotype is expressed in a heterozygous diploid cell. The diploid has the same phenotype as the wild type Recessive: wild type phenotype MUT 1 mut 1 Dominant: mutant phenotype

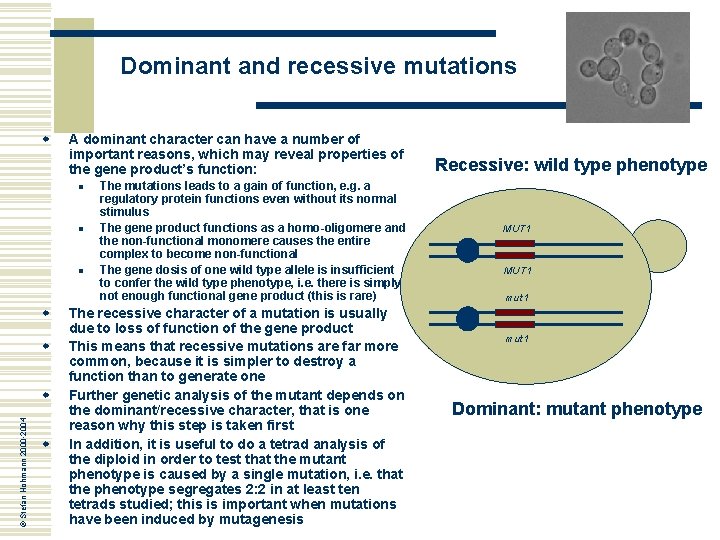
Dominant and recessive mutations w A dominant character can have a number of important reasons, which may reveal properties of the gene product’s function: n n n w w © Stefan Hohmann 2000 -2004 w w The mutations leads to a gain of function, e. g. a regulatory protein functions even without its normal stimulus The gene product functions as a homo-oligomere and the non-functional monomere causes the entire complex to become non-functional The gene dosis of one wild type allele is insufficient to confer the wild type phenotype, i. e. there is simply not enough functional gene product (this is rare) The recessive character of a mutation is usually due to loss of function of the gene product This means that recessive mutations are far more common, because it is simpler to destroy a function than to generate one Further genetic analysis of the mutant depends on the dominant/recessive character, that is one reason why this step is taken first In addition, it is useful to do a tetrad analysis of the diploid in order to test that the mutant phenotype is caused by a single mutation, i. e. that the phenotype segregates 2: 2 in at least ten tetrads studied; this is important when mutations have been induced by mutagenesis Recessive: wild type phenotype MUT 1 mut 1 Dominant: mutant phenotype

Yeast genetics: markers © Stefan Hohmann 2000 -2004 Nella pratica industriale i markers per l’auxotrofia non sono consigliati

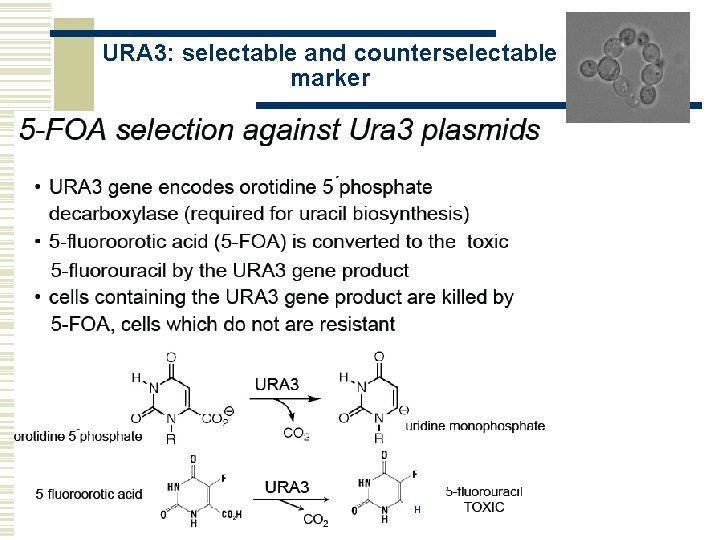
© Stefan Hohmann 2000 -2004 URA 3: selectable and counterselectable marker H

© Stefan Hohmann 2000 -2004 Cloning in yeast: plasmids classification

Cloning in yeast w w © Stefan Hohmann 2000 -2004 w w The era of yeast molecular genetics started as early as 1978, when S. cerevisiae was first transformed successfully with foreign DNA There are numerous transformation protocols but all are at least three orders of magnitude less efficient as transformation in E. coli Yeast can maintain replicating plasmids but the copy number is much smaller than in E. coli, usually between one and 50 per cell Yeast can maintain more than one type of plasmid at the same time. This can complicate gene cloning from a library. It can also be very useful to transform yeast with two different plasmids simultaneously, for instance for a method called plasmid shuffling Cloning and plasmid preparation from yeast is very ineffective Therefore, cloning in yeast uses E. coli as a plasmid production system: n n Plasmids are constructed in vitro Plasmids are transformed into E. coli and the constructions are confirmed, just in the same way as when working with bacteria Plasmids are produced in bacteria. . . . and then transformed into yeast Hence we work with so-called yeast-E. coli shuttle vectors On the other hand, yeast has a very efficient and reliable system for homologous recombination, which can be used for cloning

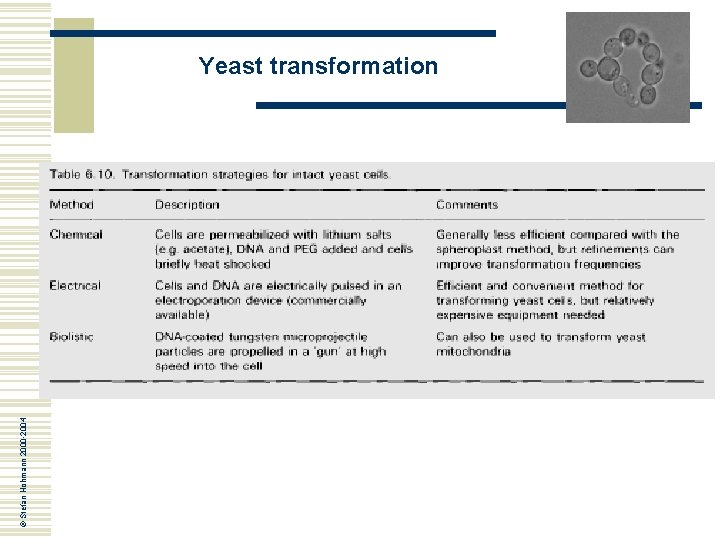
© Stefan Hohmann 2000 -2004 Yeast transformation

© Stefan Hohmann 2000 -2004 Cloning in yeast: plasmids classification

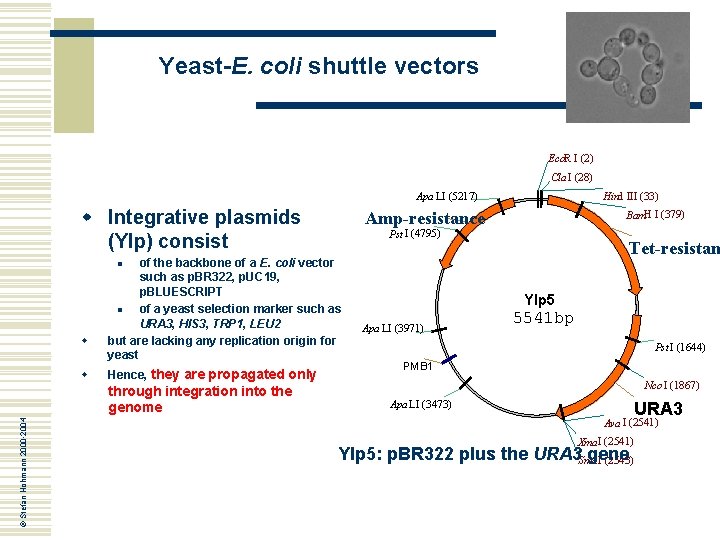
Yeast-E. coli shuttle vectors Eco. R I (2) Cla I (28) Apa LI (5217) w Integrative plasmids (YIp) consist Hind III (33) Amp-resistance Bam. H I (379) Pst I (4795) of the backbone of a E. coli vector such as p. BR 322, p. UC 19, p. BLUESCRIPT n of a yeast selection marker such as URA 3, HIS 3, TRP 1, LEU 2 but are lacking any replication origin for yeast Tet-resistan n w w Hence, they are propagated only © Stefan Hohmann 2000 -2004 through integration into the genome YIp 5 Apa LI (3971) 5541 bp Pst I (1644) PMB 1 Nco I (1867) Apa LI (3473) URA 3 Ava I (2541) Xma I (2541) YIp 5: p. BR 322 plus the URA 3 Sma gene I (2543)

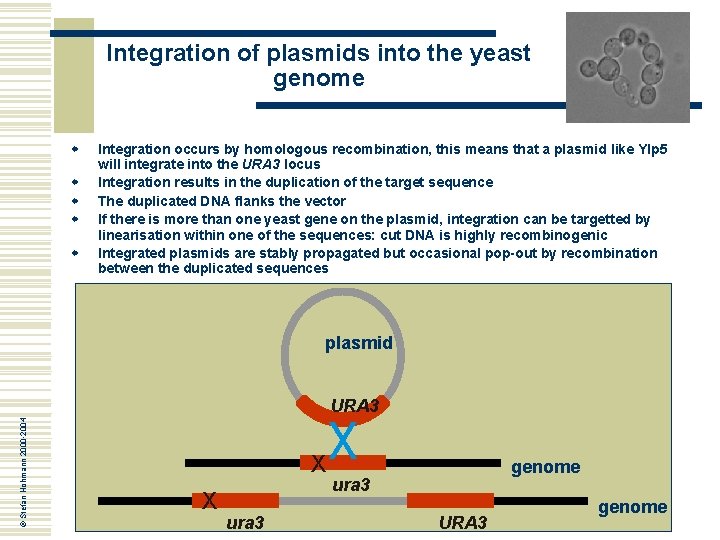
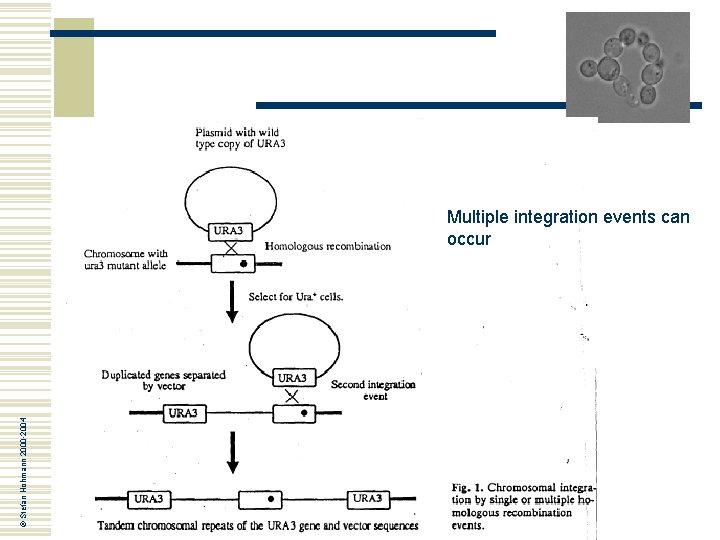
Integration of plasmids into the yeast genome w w w Integration occurs by homologous recombination, this means that a plasmid like YIp 5 will integrate into the URA 3 locus Integration results in the duplication of the target sequence The duplicated DNA flanks the vector If there is more than one yeast gene on the plasmid, integration can be targetted by linearisation within one of the sequences: cut DNA is highly recombinogenic Integrated plasmids are stably propagated but occasional pop-out by recombination between the duplicated sequences plasmid © Stefan Hohmann 2000 -2004 URA 3 X X ura 3 genome ura 3 URA 3 genome

© Stefan Hohmann 2000 -2004 Multiple integration events can occur

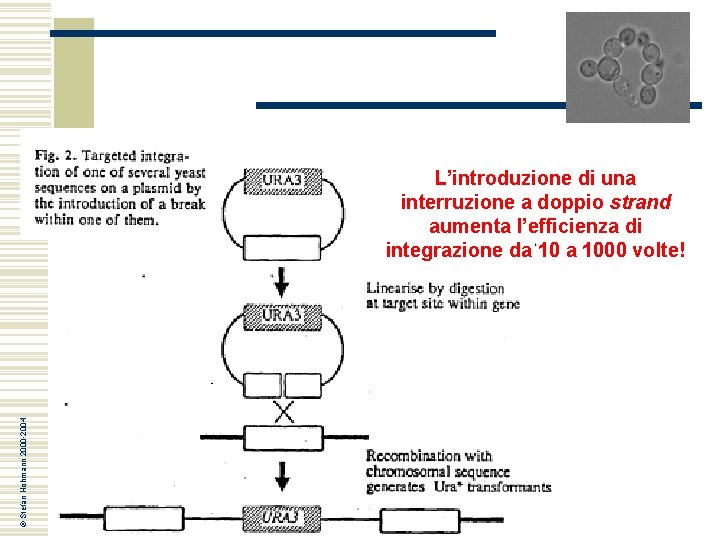
© Stefan Hohmann 2000 -2004 L’introduzione di una interruzione a doppio strand aumenta l’efficienza di integrazione da 1000 volte!

© Stefan Hohmann 2000 -2004 Cloning in yeast: plasmids classification

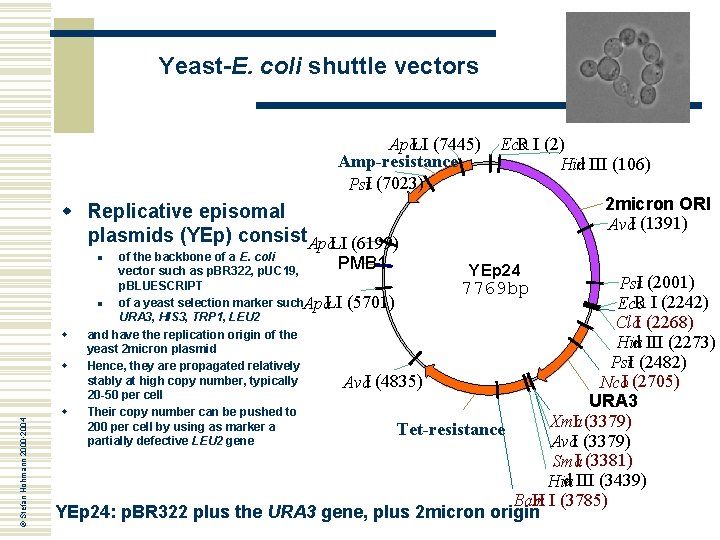
Yeast-E. coli shuttle vectors Apa. LI (7445) Amp-resistance Pst. I (7023) w Replicative episomal plasmids (YEp) consist. Apa. LI (6199) R I (2) Eco d III (106) Hin of the backbone of a E. coli PMB 1 vector such as p. BR 322, p. UC 19, YEp 24 p. BLUESCRIPT 7769 bp n of a yeast selection marker such. Apa LI (5701) URA 3, HIS 3, TRP 1, LEU 2 and have the replication origin of the yeast 2 micron plasmid Hence, they are propagated relatively stably at high copy number, typically Ava. I (4835) 20 -50 per cell Their copy number can be pushed to 200 per cell by using as marker a Tet-resistance partially defective LEU 2 gene 2 micron ORI Ava. I (1391) © Stefan Hohmann 2000 -2004 n Pst. I (2001) R I (2242) Eco Cla. I (2268) w d III (2273) Hin Pst. I (2482) w Nco. I (2705) URA 3 w I (3379) Xma Ava. I (3379) Sma. I (3381) d III (3439) Hin H I (3785) Bam YEp 24: p. BR 322 plus the URA 3 gene, plus 2 micron origin

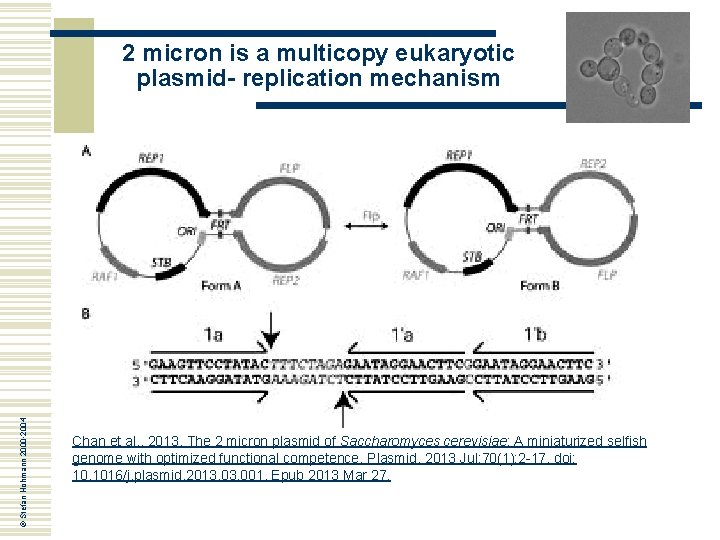
© Stefan Hohmann 2000 -2004 2 micron is a multicopy eukaryotic plasmid- replication mechanism Chan et al. , 2013. The 2 micron plasmid of Saccharomyces cerevisiae: A miniaturized selfish genome with optimized functional competence. Plasmid. 2013 Jul; 70(1): 2 -17. doi: 10. 1016/j. plasmid. 2013. 001. Epub 2013 Mar 27.

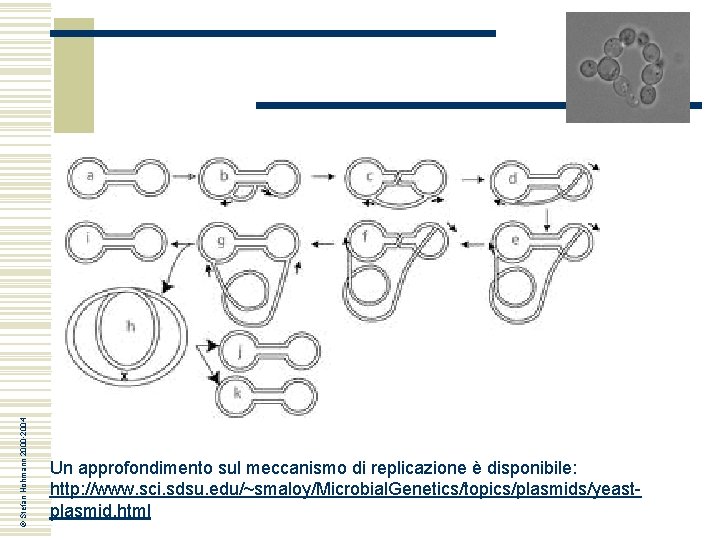
© Stefan Hohmann 2000 -2004 Un approfondimento sul meccanismo di replicazione è disponibile: http: //www. sci. sdsu. edu/~smaloy/Microbial. Genetics/topics/plasmids/yeastplasmid. html

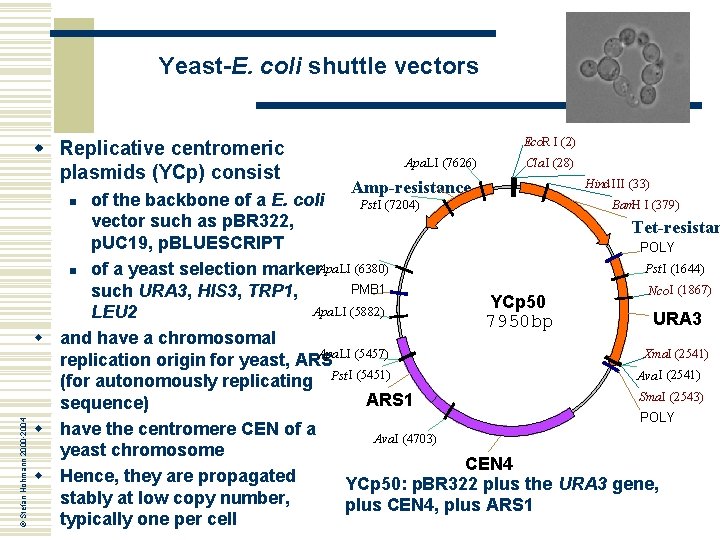
Yeast-E. coli shuttle vectors w Replicative centromeric plasmids (YCp) consist Apa LI (7626) Amp-resistance Cla I (28) Hind III (33) of the backbone of a E. coli Pst I (7204) Bam. H I (379) vector such as p. BR 322, Tet-resistan p. UC 19, p. BLUESCRIPT POLY Pst I (1644) n of a yeast selection marker. Apa LI (6380) PMB 1 Nco I (1867) such URA 3, HIS 3, TRP 1, YCp 50 Apa LI (5882) LEU 2 URA 3 7950 bp w and have a chromosomal Apa LI (5457) Xma. I (2541) replication origin for yeast, ARS Ava I (2541) (for autonomously replicating Pst I (5451) Sma. I (2543) ARS 1 sequence) POLY w have the centromere CEN of a Ava I (4703) yeast chromosome CEN 4 w Hence, they are propagated YCp 50: p. BR 322 plus the URA 3 gene, stably at low copy number, plus CEN 4, plus ARS 1 typically one per cell n © Stefan Hohmann 2000 -2004 Eco. R I (2)

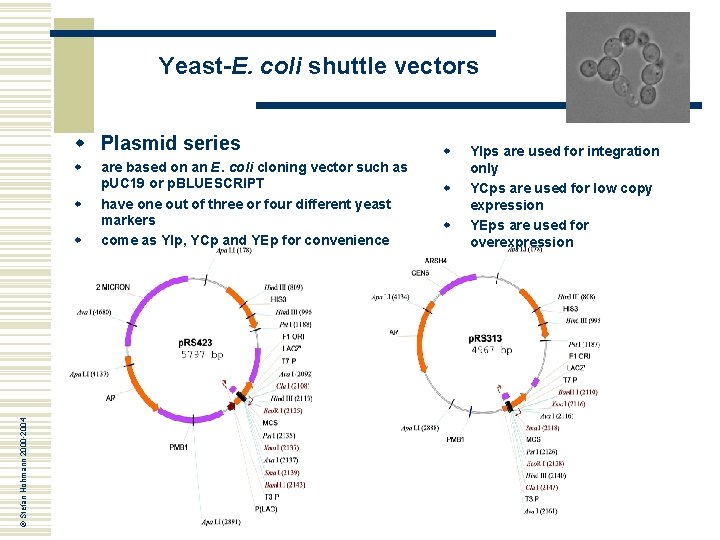
Yeast-E. coli shuttle vectors w Plasmid series w w © Stefan Hohmann 2000 -2004 w are based on an E. coli cloning vector such as p. UC 19 or p. BLUESCRIPT have one out of three or four different yeast markers come as YIp, YCp and YEp for convenience w w w YIps are used for integration only YCps are used for low copy expression YEps are used for overexpression

© Stefan Hohmann 2000 -2004 Cloning in yeast: plasmids classification

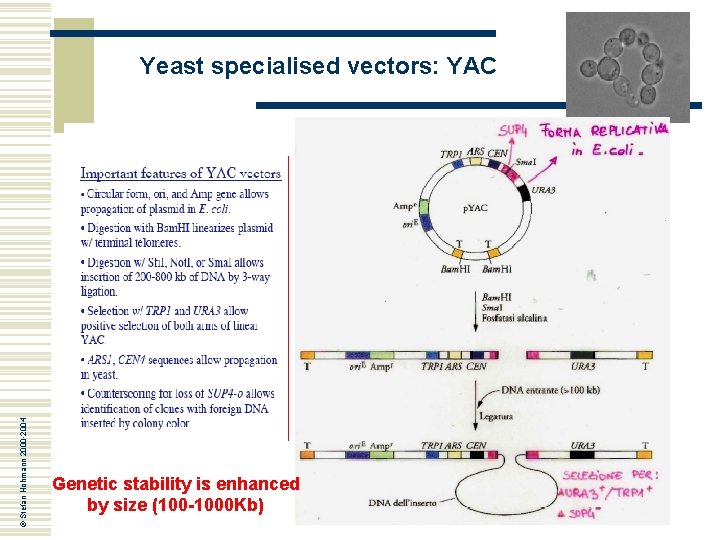
© Stefan Hohmann 2000 -2004 Yeast specialised vectors: YAC Genetic stability is enhanced by size (100 -1000 Kb)

Yeast specialised vectors: YAC Human genome project © Stefan Hohmann 2000 -2004 3, 234. 83 Mb (Mega-basepairs) per haploid genome This is a photo of two copies of the Washington University Human Genome YAC Library. Each of the stacks is approximately 12 microtiter plates. Each plate has 96 wells, each with different yeast clones.

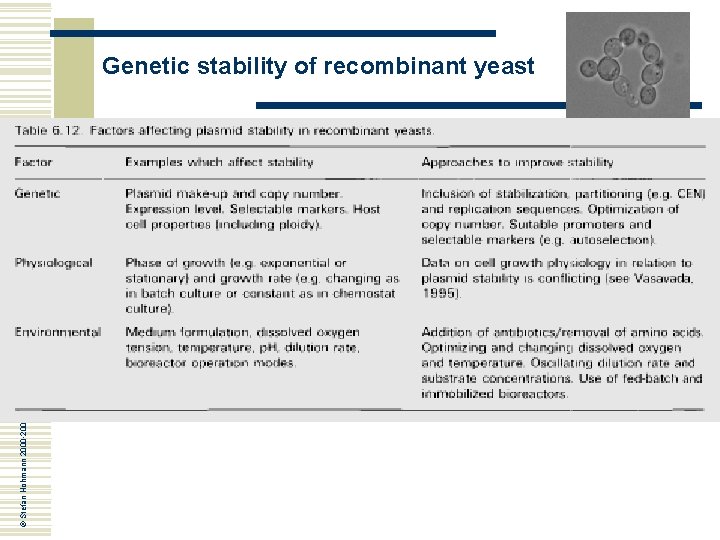
© Stefan Hohmann 2000 -2004 Genetic stability of recombinant yeast