Yeast Comparative Genomic Hybridization CGH A method for
Yeast Comparative Genomic Hybridization (CGH): A method for microarray detection of aneuploidy in S. cerevisiae Jackie Ryan Honors Thesis Defense April 20, 2006
Overview • DNA Microarrays – Typical Use – CGH arrays • Development of CGH Procedure – Basic Method – Optimization – Validation • Future use of CGH Procedure at Davidson
Typical DNA Microarray
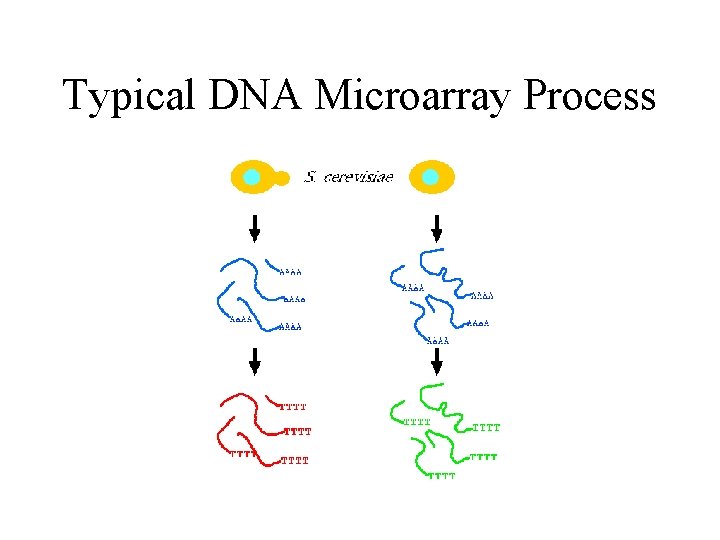
Typical DNA Microarray Process
Typical DNA Microarray Process

Typical DNA Microarray Process
Yeast c. DNA Microarray
CGH: An Alternative Use of DNA Microarrays • Hybridize genomic DNA to array • Detect deletions/amplifications of genes (aneuploidy) • Applications: – Laboratory Evolution – Cancer

Laboratory Evolution • � Stressed yeast = aneuploidy – Acetone & Cold, Glucose-limited conditions • Under glucose-limited conditions – Chromosomal rearrangements (Dunham et al. , 2002) – Abnormal copy number of genes (Ferea et al. , 1999)
Human Cancers • Aneuploidy and disease • Cancer – Cell division pathways • BUB 1 B – Multiple hit hypothesis • Oncogenes • Tumor-suppressors

Questions to Answer • Is aneuploidy random? • Is it reproducible? – Position – Sequence – Function
Advantages of CGH • High-throughput – Identify: candidate genes, patterns • Compare two different populations – wild type vs. evolved – normal tissue vs. cancerous tissue
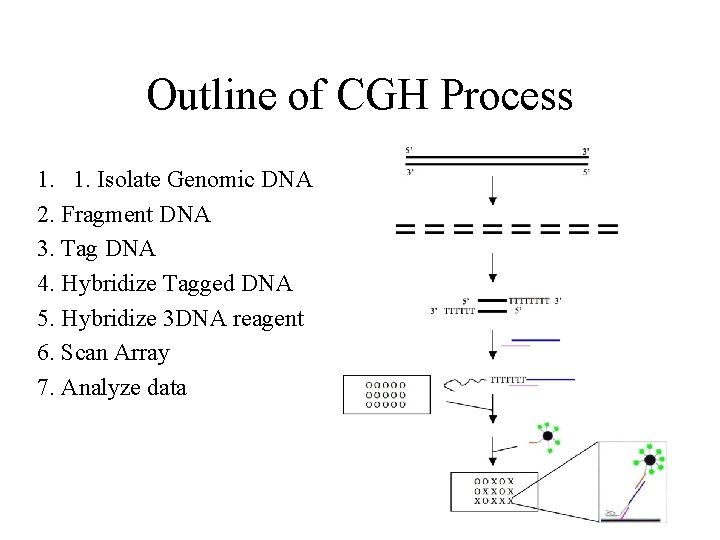
Outline of CGH Process 1. 1. Isolate Genomic DNA 2. Fragment DNA 3. Tag DNA 4. Hybridize Tagged DNA 5. Hybridize 3 DNA reagent 6. Scan Array 7. Analyze data
Tagging Method • Genisphere 3 DNA Array 900 DNA kit Alexa 546/Alexa 647 – Robust Signal – Less photobleaching
Hypothetical CGH Experiment Red = wt Green = Evolved
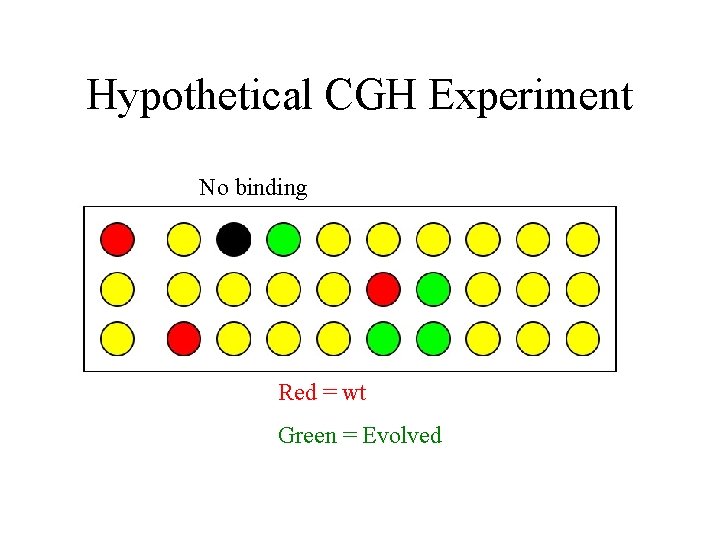
Hypothetical CGH Experiment No binding Red = wt Green = Evolved
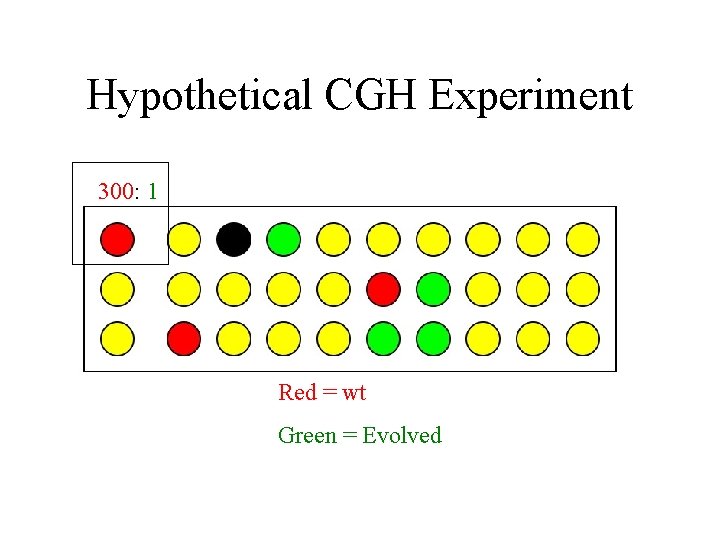
Hypothetical CGH Experiment 300: 1 Red = wt Green = Evolved
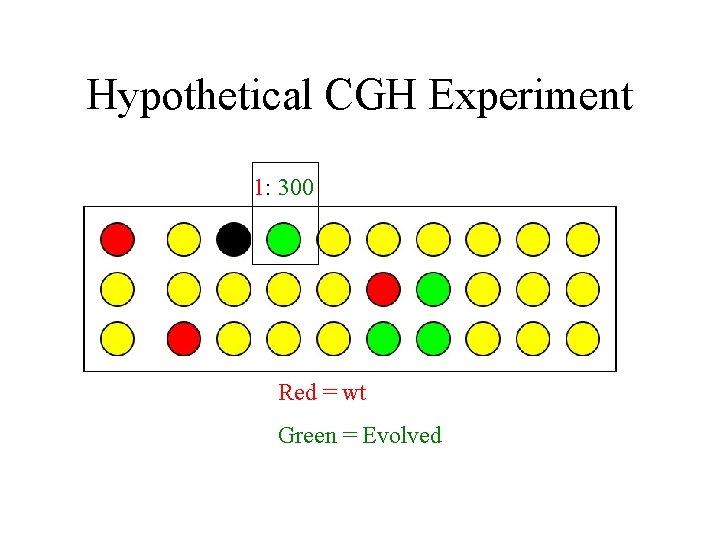
Hypothetical CGH Experiment 1: 300 Red = wt Green = Evolved
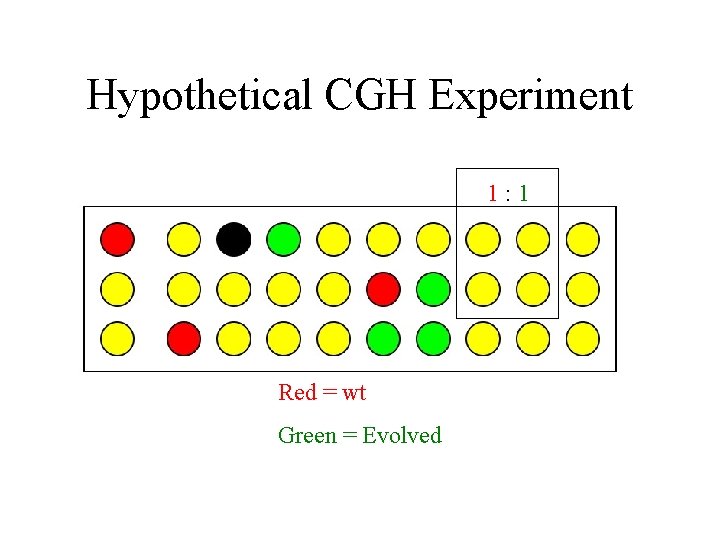
Hypothetical CGH Experiment 1: 1 Red = wt Green = Evolved
Genomic Isolation Method • Factors Considered – Toxicity – Time – Cost – Ease of use • Zymo kits
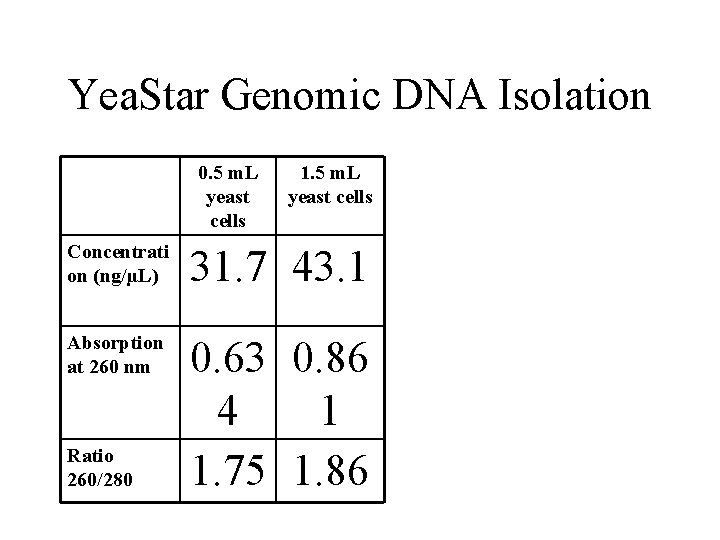
Yea. Star Genomic DNA Isolation 0. 5 m. L yeast cells 1. 5 m. L yeast cells Concentrati on (ng/µL) 31. 7 43. 1 Absorption at 260 nm 0. 63 0. 86 4 1 1. 75 1. 86 Ratio 260/280
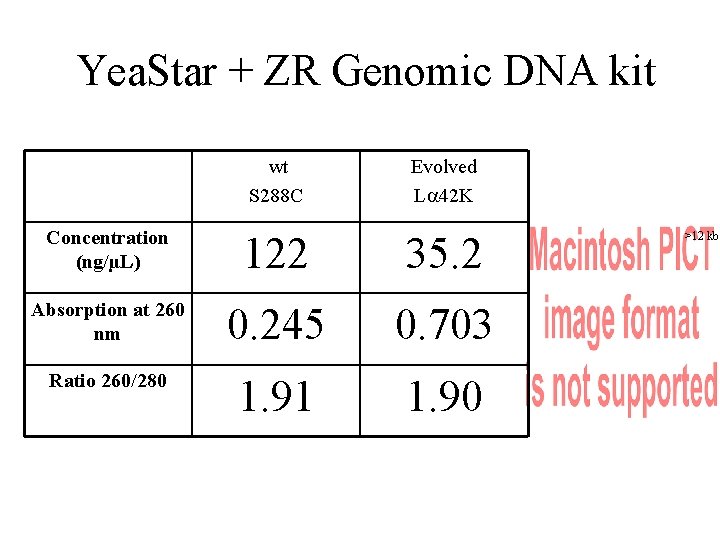
Yea. Star + ZR Genomic DNA kit wt S 288 C Evolved L 42 K Concentration (ng/µL) 122 35. 2 Absorption at 260 nm 0. 245 0. 703 Ratio 260/280 1. 91 1. 90 >12 kb
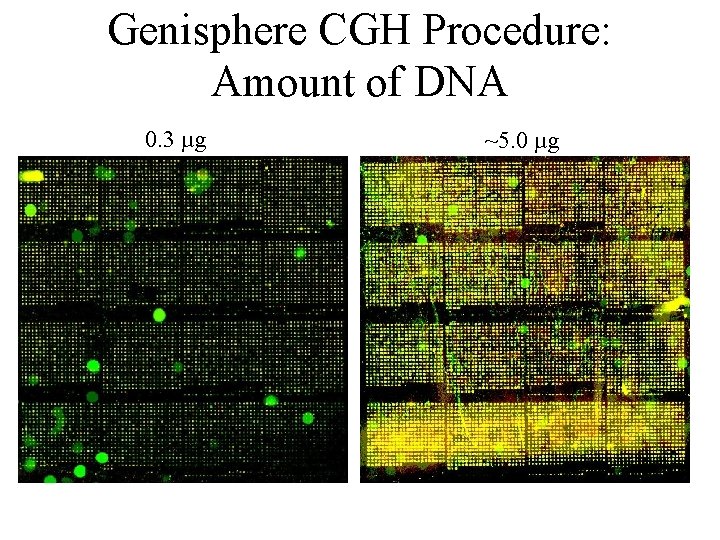
Genisphere CGH Procedure: Amount of DNA 0. 3 µg ~5. 0 µg
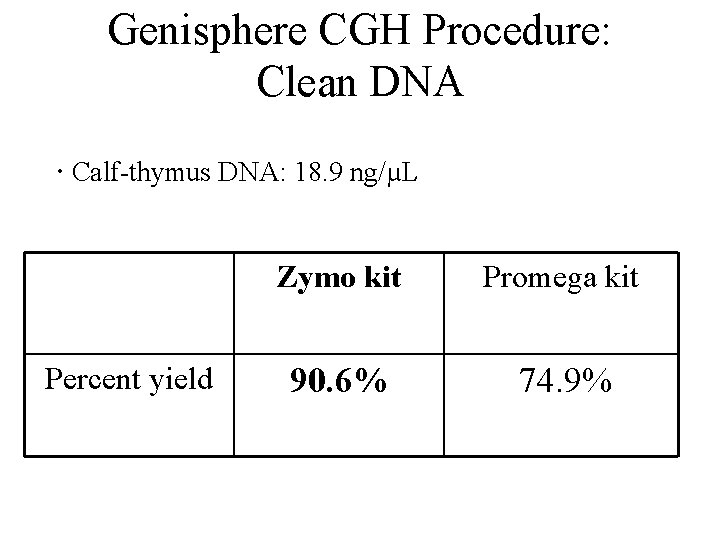
Genisphere CGH Procedure: Clean DNA · Calf-thymus DNA: 18. 9 ng/µL Percent yield Zymo kit Promega kit 90. 6% 74. 9%
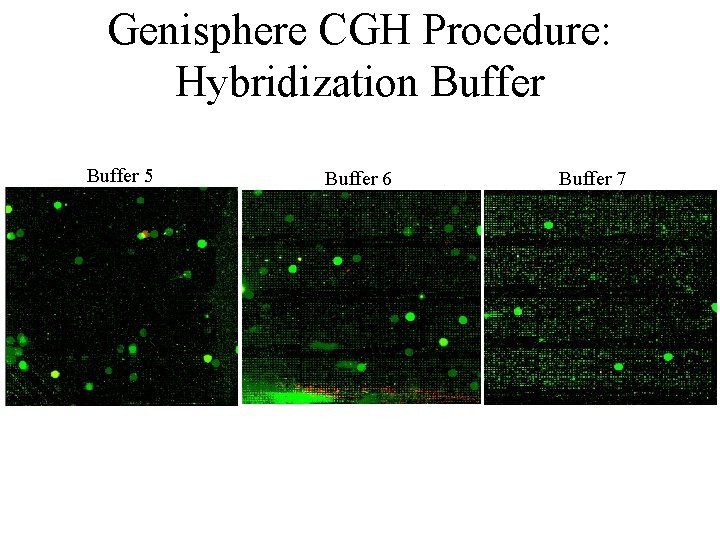
Genisphere CGH Procedure: Hybridization Buffer 5 Buffer 6 Buffer 7
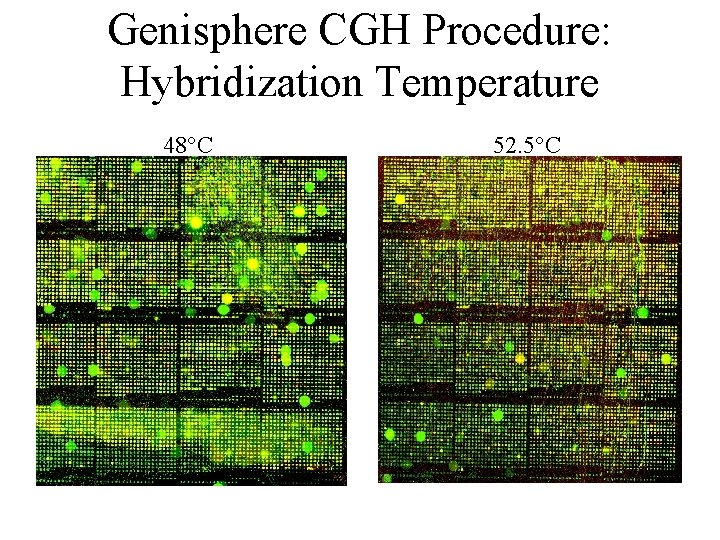
Genisphere CGH Procedure: Hybridization Temperature 48°C 52. 5°C
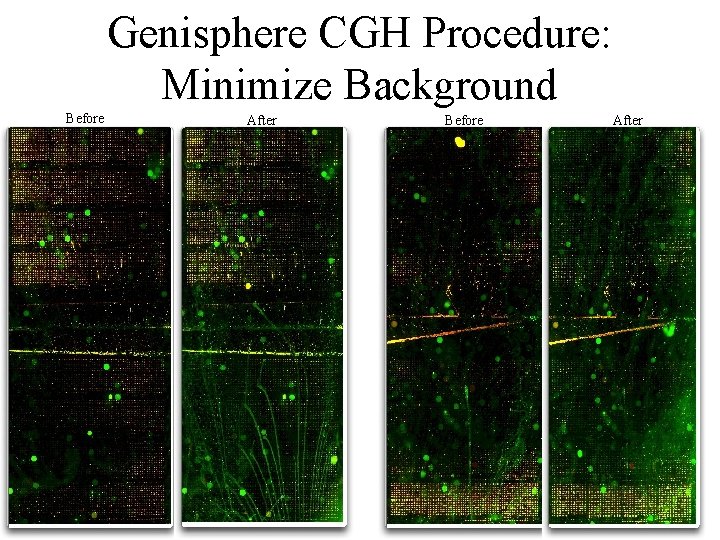
Genisphere CGH Procedure: Minimize Background Before After
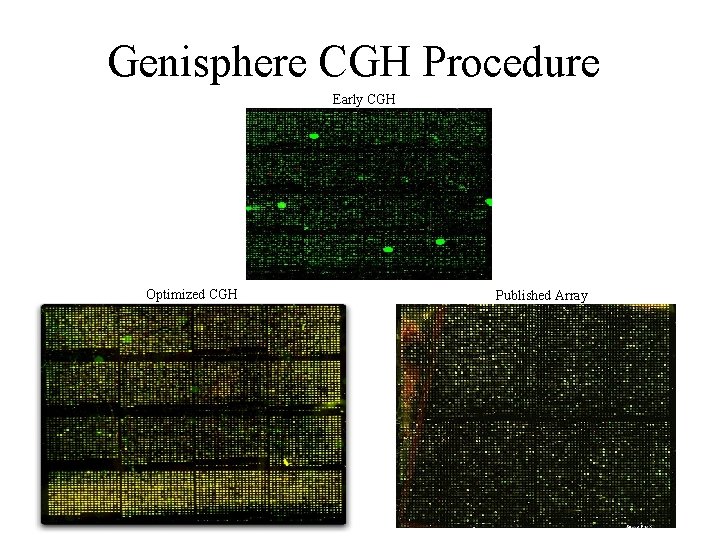
Genisphere CGH Procedure Early CGH Optimized CGH Published Array
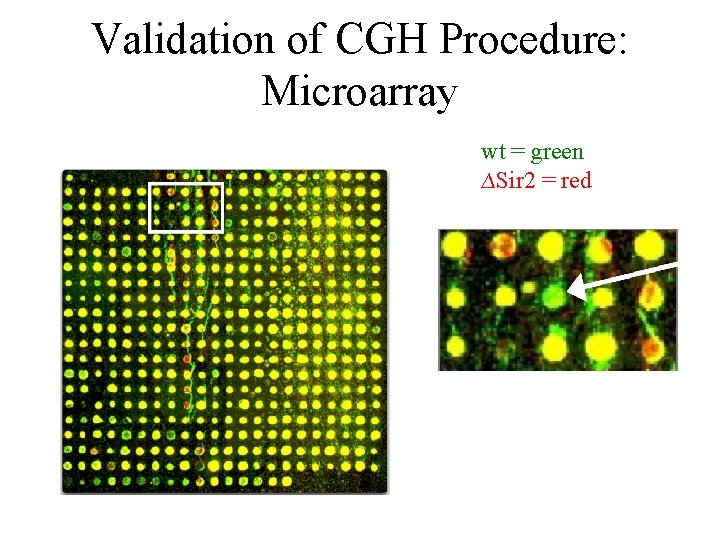
Validation of CGH Procedure: Microarray wt = green ∆Sir 2 = red
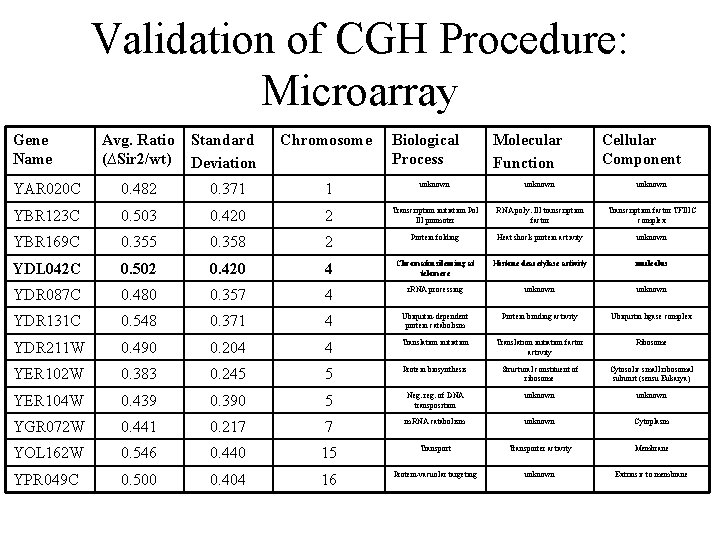
Validation of CGH Procedure: Microarray Gene Name Avg. Ratio (∆Sir 2/wt) Standard Deviation Chromosome Biological Process Molecular Function Cellular Component YAR 020 C 0. 482 0. 371 1 unknown YBR 123 C 0. 503 0. 420 2 Transcription initiation Pol III promoter RNA poly. III transcription factor TFIIIC complex YBR 169 C 0. 355 0. 358 2 Protein folding Heat shock protein activity unknown YDL 042 C 0. 502 0. 420 4 Chromatin silencing at telomere Histone deacetylase activity nucleolus YDR 087 C 0. 480 0. 357 4 r. RNA processing unknown YDR 131 C 0. 548 0. 371 4 Ubiquitin-dependent protein catabolism Protein binding activity Ubiquitin ligase complex YDR 211 W 0. 490 0. 204 4 Translation initiation factor activity Ribosome YER 102 W 0. 383 0. 245 5 Protein biosynthesis Structural constituent of ribosome Cytosolic small ribosomal subunit (sensu Eukarya) YER 104 W 0. 439 0. 390 5 Neg. reg. of DNA transposition unknown YGR 072 W 0. 441 0. 217 7 m. RNA catabolism unknown Cytoplasm YOL 162 W 0. 546 0. 440 15 Transporter activity Membrane YPR 049 C 0. 500 0. 404 16 Protein-vacuolar targeting unknown Extrinsic to membrane
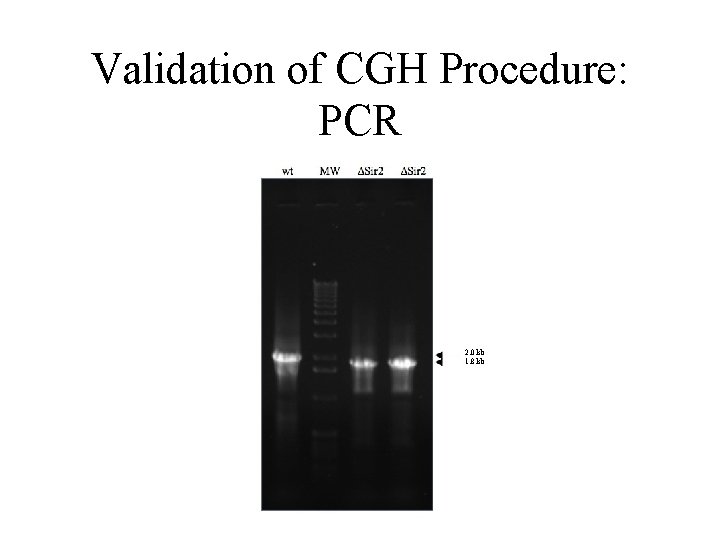
Validation of CGH Procedure: PCR 2. 0 kb 1. 8 kb
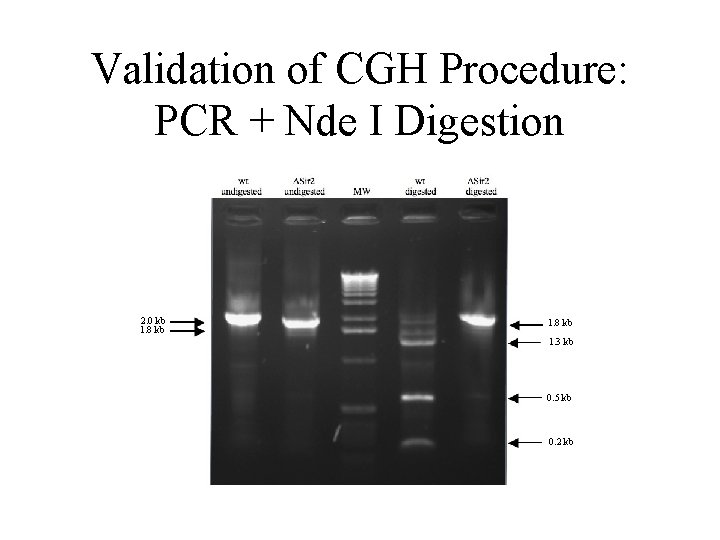
Validation of CGH Procedure: PCR + Nde I Digestion 2. 0 kb 1. 8 kb 1. 3 kb 0. 5 kb 0. 2 kb

False Positives · Self vs. Self Arrays visually show consistent green and red spots

False Positives · Compile numerical data - 51 spots were consistently green or red · Hypothesis: 3 DNA reagent bind directly to spots · LALIGN to test
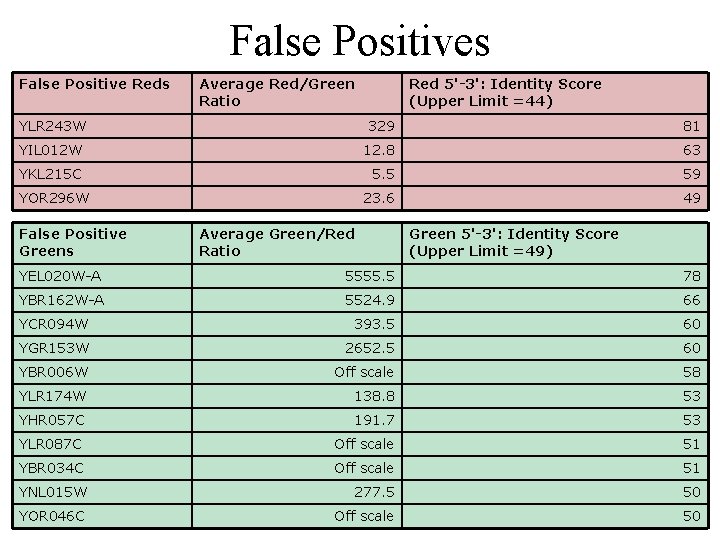
False Positives False Positive Reds Average Red/Green Ratio Red 5'-3': Identity Score (Upper Limit =44) YLR 243 W 329 81 YIL 012 W 12. 8 63 YKL 215 C 5. 5 59 23. 6 49 YOR 296 W False Positive Greens Average Green/Red Ratio Green 5'-3': Identity Score (Upper Limit =49) YEL 020 W-A 5555. 5 78 YBR 162 W-A 5524. 9 66 YCR 094 W 393. 5 60 YGR 153 W 2652. 5 60 YBR 006 W Off scale 58 YLR 174 W 138. 8 53 YHR 057 C 191. 7 53 YLR 087 C Off scale 51 YBR 034 C Off scale 51 YNL 015 W 277. 5 50 YOR 046 C Off scale 50

Future Work with CGH Procedure • Mutant Yeast from Dr. Clifford Zeyl – Evolution under glucose-limited conditions • 2, 000 generations • 5, 000 generations

Acknowledgments • • Davidson College – Biology Department – Dr. Malcolm Campbell – Dr. Laurie Heyer – Dr. Karen Bernd – Chris Healey – Peggy Maiorano – Emily Oldham and previous genomics students – Lab mates: Matt Gemberling, Mac Cowell, Kristen De. Celle, Franois Trappey, Andrew Drysdale, and Oscar Hernandez Other Institutions – Dr. Todd Eckdahl of Missouri Western State College – Dr. Laura Hoopes of Pomona College – Dr. Clifford Zeyl of Wake Forest University – GCAT – Genisphere and Zymo Research
- Slides: 37