Workshop Analysis of Microbial Transcriptomics Data Data Analysis
Workshop: Analysis of Microbial Transcriptomics Data - Data Analysis of m. RNA expression profiles - Identifiaction of differential gene-expression Martien Caspers, BSc Remco Kort, Prof, Dr TNO, Microbiology & Systems Biology Zeist, NL 16 sep 2011, VU, Amsterdam

m. RNA expression profiles • Arrays (this course): – Hybridisation of (fluorescently) labelled c. DNA populations (whole cell) to arrays with probes of known DNA sequences / functions / genes – Result: Signal/probe • RNAseq: – Mass-sequencing of c. DNA populations – Mass-Blast to annotated genome(s) – Result: nr of sequences per gene / genetic-element
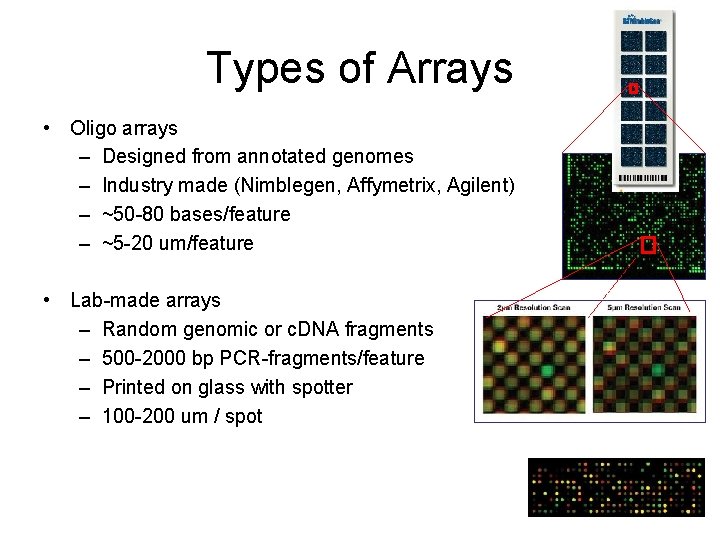
Types of Arrays • Oligo arrays – Designed from annotated genomes – Industry made (Nimblegen, Affymetrix, Agilent) – ~50 -80 bases/feature – ~5 -20 um/feature • Lab-made arrays – Random genomic or c. DNA fragments – 500 -2000 bp PCR-fragments/feature – Printed on glass with spotter – 100 -200 um / spot
Experimental use of arrays • Double hybridisation: 2 color – Treated Sample e. g. Cy 5 -labelled – Untreated sample (ctrl) e. g. Cy 3 -labelled – Each spot it’s own control correction of spot differences/slide needed for lab spotted arrays – Disadvantage: each slide needs ctrl • Single hybridisation (this presentation) – Industry arrays reproducible spots from slide to slide – Each sample on separate slide
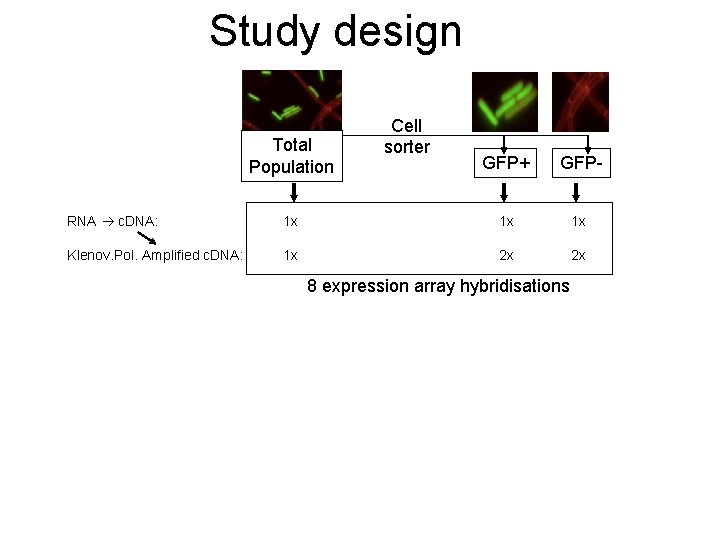
Study design Total Population Cell sorter GFP+ GFP- RNA c. DNA: 1 x 1 x 1 x Klenov. Pol. Amplified c. DNA: 1 x 2 x 2 x 8 expression array hybridisations

Public Micro. Array Data: Geo database http: //www. ncbi. nlm. nih. gov/geo/ -Study identifier: GSE 16345 -B. subtilis heterogeneity motile/non-motile
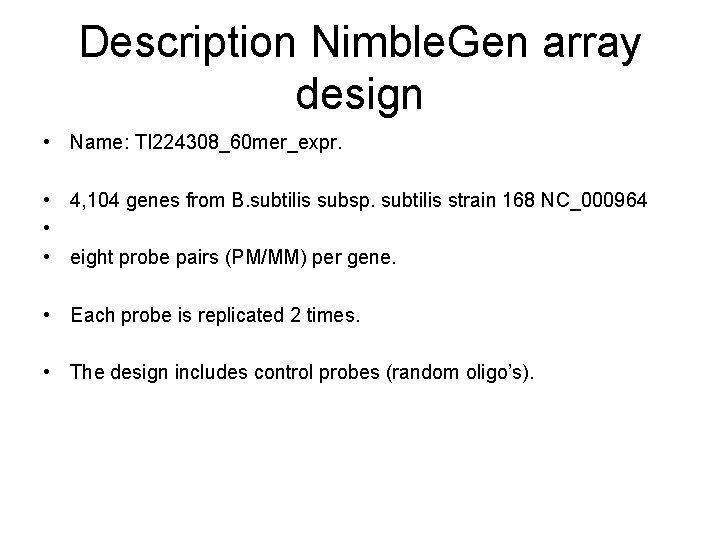
Description Nimble. Gen array design • Name: TI 224308_60 mer_expr. • 4, 104 genes from B. subtilis subsp. subtilis strain 168 NC_000964 • • eight probe pairs (PM/MM) per gene. • Each probe is replicated 2 times. • The design includes control probes (random oligo’s).

Study design Total Population Cell sorter GFP+ GFP- RNA c. DNA: 1 x 1 x 1 x Klenov. Pol. Amplified c. DNA: 1 x 2 x 2 x 8 expression array hybridisations
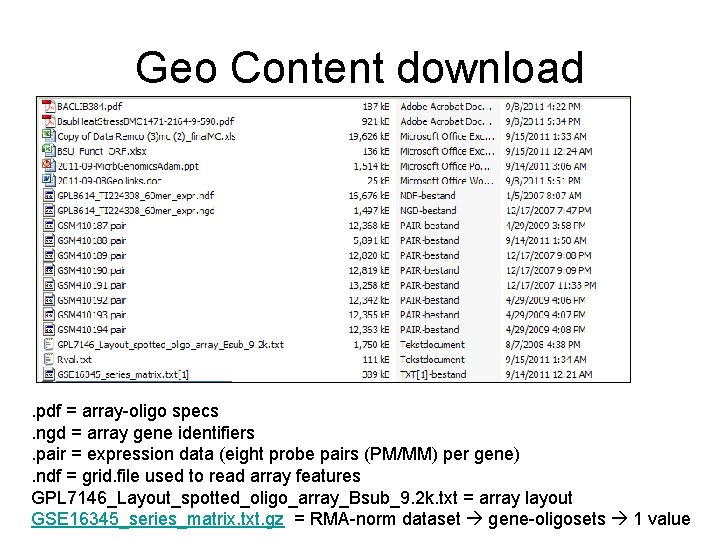
Geo Content download . pdf = array-oligo specs. ngd = array gene identifiers. pair = expression data (eight probe pairs (PM/MM) per gene). ndf = grid. file used to read array features GPL 7146_Layout_spotted_oligo_array_Bsub_9. 2 k. txt = array layout GSE 16345_series_matrix. txt. gz = RMA-norm dataset gene-oligosets 1 value
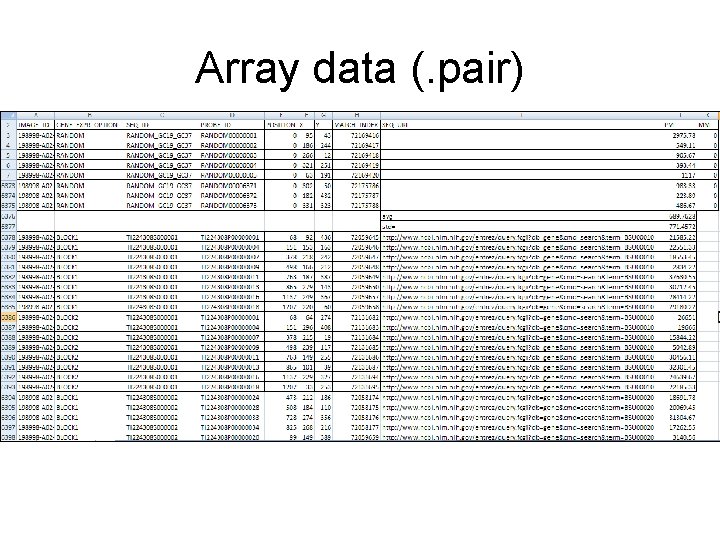
Array data (. pair)
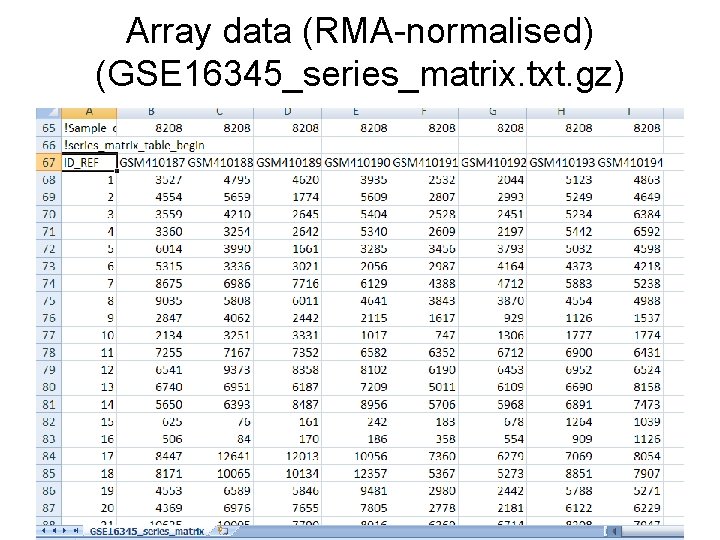
Array data (RMA-normalised) (GSE 16345_series_matrix. txt. gz)
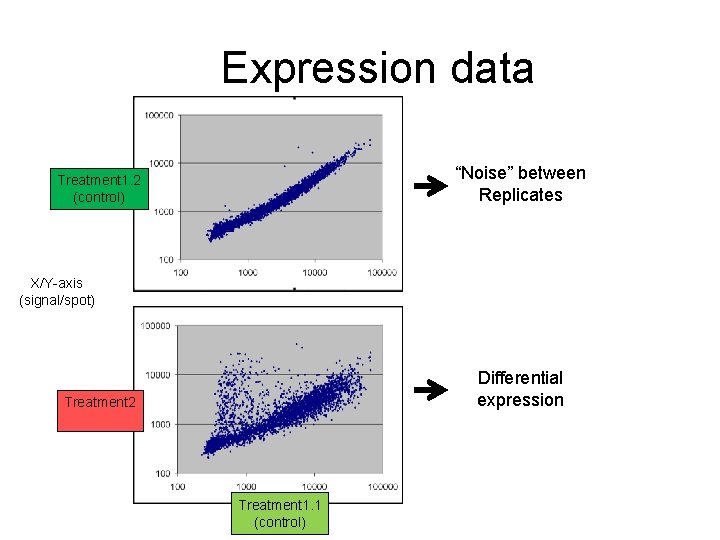
Expression data “Noise” between Replicates Treatment 1. 2 (control) X/Y-axis (signal/spot) Differential expression Treatment 2 Treatment 1. 1 (control)
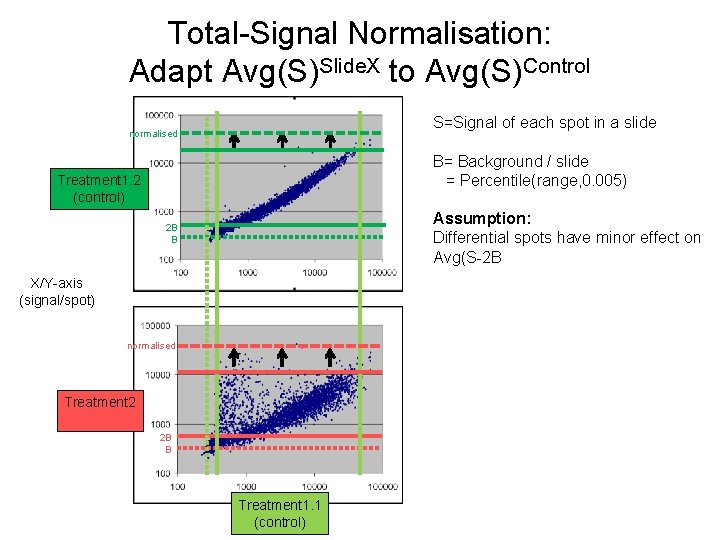
Total-Signal Normalisation: Adapt Avg(S)Slide. X to Avg(S)Control S=Signal of each spot in a slide normalised B= Background / slide = Percentile(range, 0. 005) Treatment 1. 2 (control) Assumption: Differential spots have minor effect on Avg(S-2 B 2 B B X/Y-axis (signal/spot) normalised Treatment 2 2 B B Treatment 1. 1 (control)
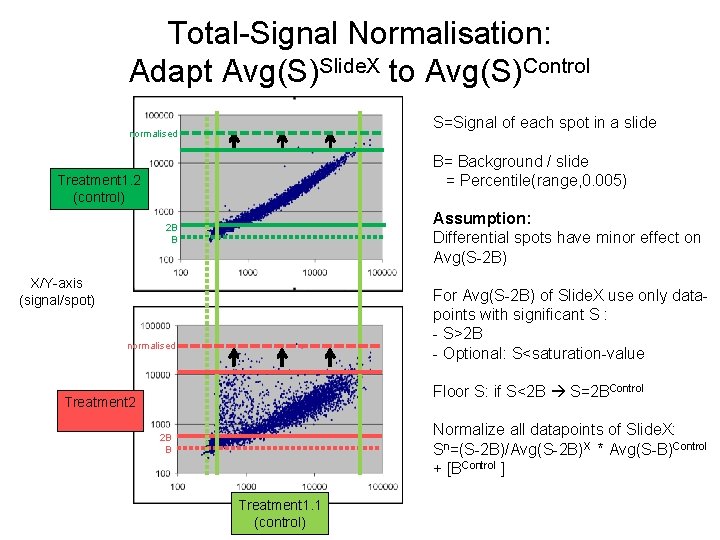
Total-Signal Normalisation: Adapt Avg(S)Slide. X to Avg(S)Control S=Signal of each spot in a slide normalised B= Background / slide = Percentile(range, 0. 005) Treatment 1. 2 (control) Assumption: Differential spots have minor effect on Avg(S-2 B) 2 B B X/Y-axis (signal/spot) For Avg(S-2 B) of Slide. X use only datapoints with significant S : - S>2 B - Optional: S<saturation-value normalised Floor S: if S<2 B S=2 BControl Treatment 2 Normalize all datapoints of Slide. X: Sn=(S-2 B)/Avg(S-2 B)X * Avg(S-B)Control + [BControl ] 2 B B Treatment 1. 1 (control)
Normalisation and Ratiocalculation in Excel • • Combine duplicate gene-probeset-data Add ORF names Add array Gene identifyers Normalise using – Avg(S) of slide “Tot. No. Amp” and Avg(S) of all other slides – 2 B (Backgr) of each slide • Calculate 2 log(R) for relevant slide pairs (e. g. GFP + / -) – R/spot = S 1/S 2 for slide 1 and 2 – 2 log. R = symmetric up/down regulation • 2 log 2 = 1 • 2 log 1 = 0 • 2 log(1/2) = -1
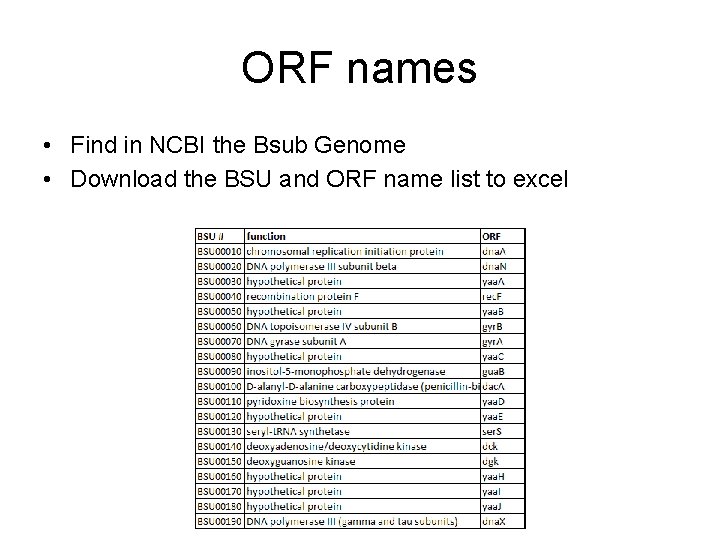
ORF names • Find in NCBI the Bsub Genome • Download the BSU and ORF name list to excel
Tprofiler: Identifies Significant Differential expression of Regulons, Pathways, Cognitive groups etc. Profilers 1) http: //www. science. uva. nl/~boorsma/t-profiler-bacillusnew/ (needs ORF + R-column) 2) http: //biocyc. org/LMON 265669/expression. html 3) http: //mgv 2. cmbi. ru. nl/genome/index. html 4) http: //www. geneontology. org/ Try First
Workshop Task • Identify Genes/Paths/Regulons related with motility related GFPexpression • Get data from internet (Geo, NCBI …. ? ) • Pre-process data in Excel (normalisation etc. ) • Identify relevant scientific questions and corresponding array-pair comparisons and controls, e. g. : – +/- GFP – Reproducibility – Effect of c. DNA-amplification
- Slides: 18