Workshop Agenda Model Overview Model history and features




- Slides: 55

Workshop Agenda Model Overview Model history and features Computational method Trajectories versus concentration Code installation Model operation Example calculations Updating HYSPLIT Meteorological Data requirements Forecast data FTP access Analysis data FTP access Display grid domain Vertical profile Contour data Examples 1 -5 Particle Trajectory Methods Trajectory computational method Trajectory example calculation trajectory model configuration Trajectory error Multiple trajectories Terrain height Meteorological analysis along a trajectory Vertical motion options PC-HYSPLIT WORKSHOP Pollutant Plume Simulations Modeling particles or puffs Concentration prediction equations Turbulence equations Dispersion model configuration Defining multiple sources Simulations using emission grids Concentration and particle display options Converting concentration data to text files Example local scale dispersion calculation Special Topics Automated trajectory calculations Trajectory cluster analysis Concentration ensembles Chemistry conversion modules Pollutant deposition Source attribution using back trajectory analysis Source attribution using source-receptor matrices Source attribution functions GIS shapefile output KML/KMZ output Customizing map labels Scripting for automated operations Extra Topics Modeling PM 10 emissions from dust storms Restarting the model from a particle dump file 116

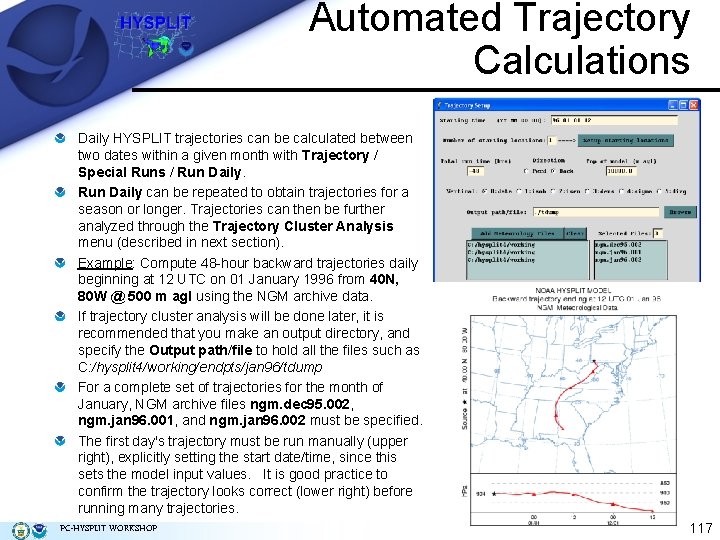
Automated Trajectory Calculations Daily HYSPLIT trajectories can be calculated between two dates within a given month with Trajectory / Special Runs / Run Daily can be repeated to obtain trajectories for a season or longer. Trajectories can then be further analyzed through the Trajectory Cluster Analysis menu (described in next section). Example: Compute 48 -hour backward trajectories daily beginning at 12 UTC on 01 January 1996 from 40 N, 80 W @ 500 m agl using the NGM archive data. If trajectory cluster analysis will be done later, it is recommended that you make an output directory, and specify the Output path/file to hold all the files such as C: /hysplit 4/working/endpts/jan 96/tdump For a complete set of trajectories for the month of January, NGM archive files ngm. dec 95. 002, ngm. jan 96. 001, and ngm. jan 96. 002 must be specified. The first day's trajectory must be run manually (upper right), explicitly setting the start date/time, since this sets the model input values. It is good practice to confirm the trajectory looks correct (lower right) before running many trajectories. PC-HYSPLIT WORKSHOP 117

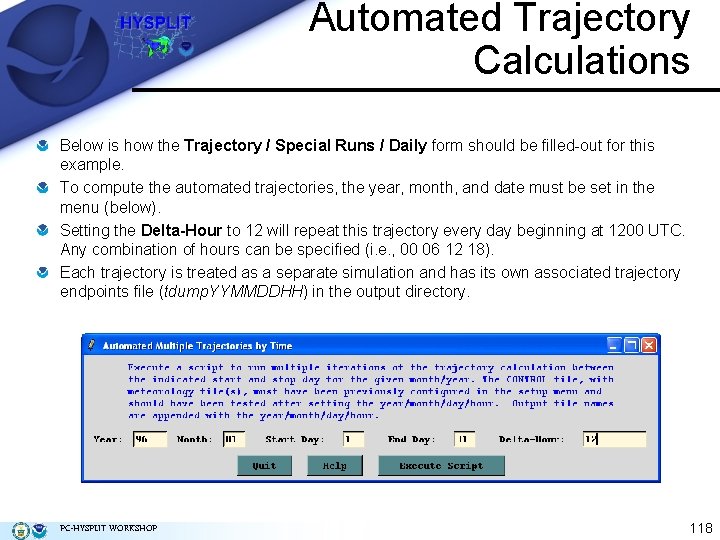
Automated Trajectory Calculations Below is how the Trajectory / Special Runs / Daily form should be filled-out for this example. To compute the automated trajectories, the year, month, and date must be set in the menu (below). Setting the Delta-Hour to 12 will repeat this trajectory every day beginning at 1200 UTC. Any combination of hours can be specified (i. e. , 00 06 12 18). Each trajectory is treated as a separate simulation and has its own associated trajectory endpoints file (tdump. YYMMDDHH) in the output directory. PC-HYSPLIT WORKSHOP 118

Trajectory Cluster Analysis is a process of grouping similar trajectories together whereby differences among individual trajectories in a cluster are minimized and differences among clusters are maximized. Ideally, each cluster represents different classes of synoptic regimes over the duration of the trajectories. Cluster analysis can be useful for matching air quality measurements with pollutant source regions. The set of January 1996 trajectories created by Run Daily in the last section will be used to illustrate the technique. The cluster analysis routine requires that the trajectory endpoints files to be used in the calculation be located in the hysplit 4clusterendpts directory. First, manually move or copy the 31 daily trajectory endpoints files from hysplit 4working to hysplit 4clusterendpts. The files from Run Daily were named tdump 96010112 (tdump. YYMMDDHH) for the 01 January trajectory to tdump 96013112 for 31 January trajectory. PC-HYSPLIT WORKSHOP 119

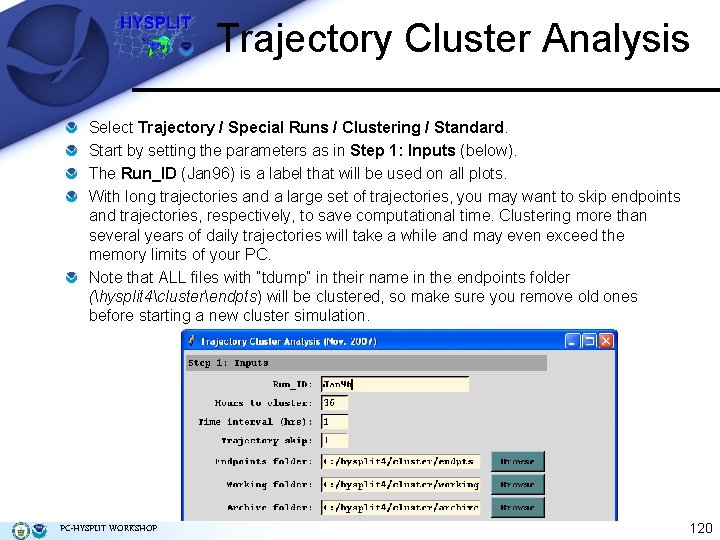
Trajectory Cluster Analysis Select Trajectory / Special Runs / Clustering / Standard. Start by setting the parameters as in Step 1: Inputs (below). The Run_ID (Jan 96) is a label that will be used on all plots. With long trajectories and a large set of trajectories, you may want to skip endpoints and trajectories, respectively, to save computational time. Clustering more than several years of daily trajectories will take a while and may even exceed the memory limits of your PC. Note that ALL files with “tdump” in their name in the endpoints folder (hysplit 4clusterendpts) will be clustered, so make sure you remove old ones before starting a new cluster simulation. PC-HYSPLIT WORKSHOP 120

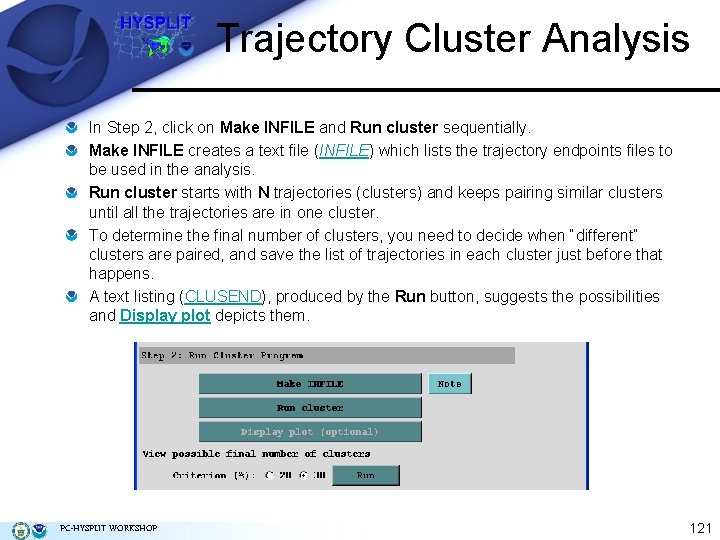
Trajectory Cluster Analysis In Step 2, click on Make INFILE and Run cluster sequentially. Make INFILE creates a text file (INFILE) which lists the trajectory endpoints files to be used in the analysis. Run cluster starts with N trajectories (clusters) and keeps pairing similar clusters until all the trajectories are in one cluster. To determine the final number of clusters, you need to decide when “different” clusters are paired, and save the list of trajectories in each cluster just before that happens. A text listing (CLUSEND), produced by the Run button, suggests the possibilities and Display plot depicts them. PC-HYSPLIT WORKSHOP 121

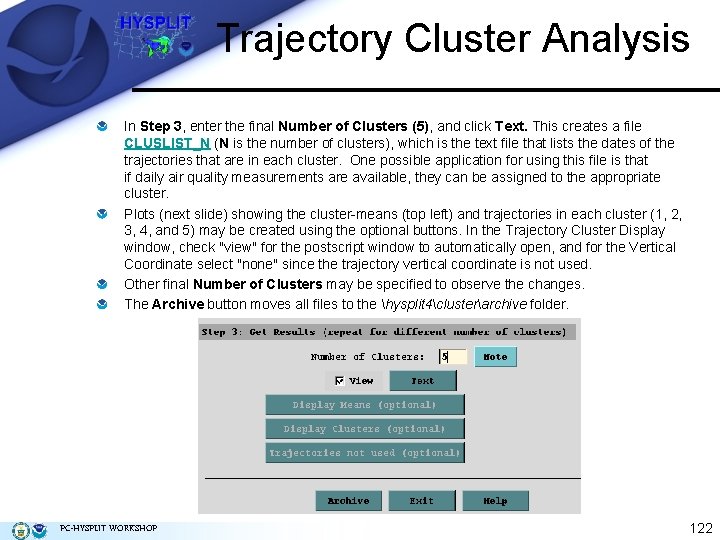
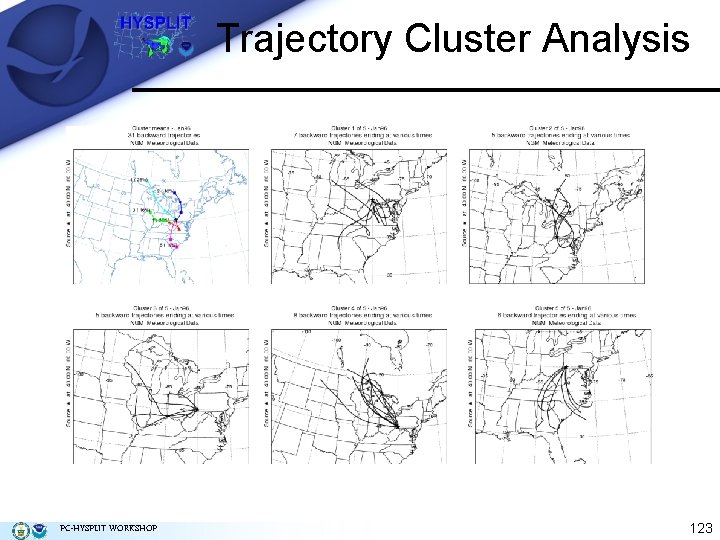
Trajectory Cluster Analysis In Step 3, enter the final Number of Clusters (5), and click Text. This creates a file CLUSLIST_N (N is the number of clusters), which is the text file that lists the dates of the trajectories that are in each cluster. One possible application for using this file is that if daily air quality measurements are available, they can be assigned to the appropriate cluster. Plots (next slide) showing the cluster-means (top left) and trajectories in each cluster (1, 2, 3, 4, and 5) may be created using the optional buttons. In the Trajectory Cluster Display window, check "view" for the postscript window to automatically open, and for the Vertical Coordinate select "none" since the trajectory vertical coordinate is not used. Other final Number of Clusters may be specified to observe the changes. The Archive button moves all files to the hysplit 4clusterarchive folder. PC-HYSPLIT WORKSHOP 122

Trajectory Cluster Analysis PC-HYSPLIT WORKSHOP 123

Concentration Ensembles • Instead of creating a single deterministic air concentration simulation, several programs are included with HYSPLIT that can be used to combine multiple HYSPLIT simulations into a single graphic that represents some variation of a concentration probability. The simplest approach is to run the model multiple times varying some parameter. • In the next example, we will run the model with several meteorological data sets, thereby varying the meteorology. We will then run the ensemble program and look at various probabilities of exceeding concentration thresholds in the results. PC-HYSPLIT WORKSHOP 124

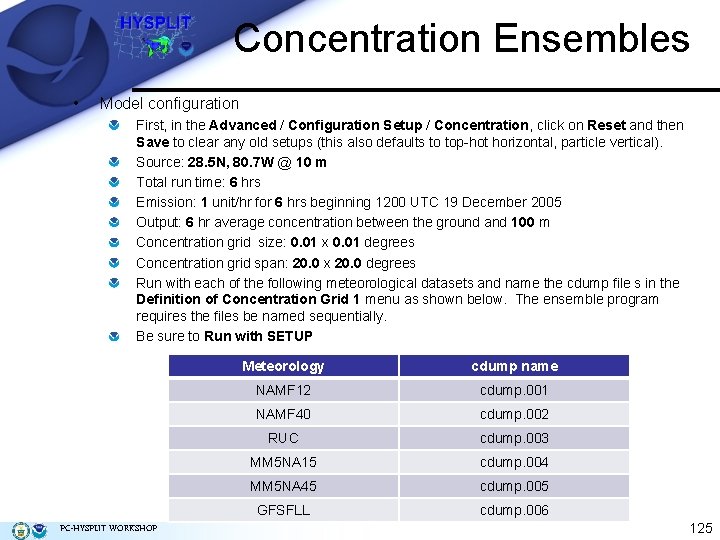
Concentration Ensembles • Model configuration First, in the Advanced / Configuration Setup / Concentration, click on Reset and then Save to clear any old setups (this also defaults to top-hot horizontal, particle vertical). Source: 28. 5 N, 80. 7 W @ 10 m Total run time: 6 hrs Emission: 1 unit/hr for 6 hrs beginning 1200 UTC 19 December 2005 Output: 6 hr average concentration between the ground and 100 m Concentration grid size: 0. 01 x 0. 01 degrees Concentration grid span: 20. 0 x 20. 0 degrees Run with each of the following meteorological datasets and name the cdump file s in the Definition of Concentration Grid 1 menu as shown below. The ensemble program requires the files be named sequentially. Be sure to Run with SETUP PC-HYSPLIT WORKSHOP Meteorology cdump name NAMF 12 cdump. 001 NAMF 40 cdump. 002 RUC cdump. 003 MM 5 NA 15 cdump. 004 MM 5 NA 45 cdump. 005 GFSFLL cdump. 006 125

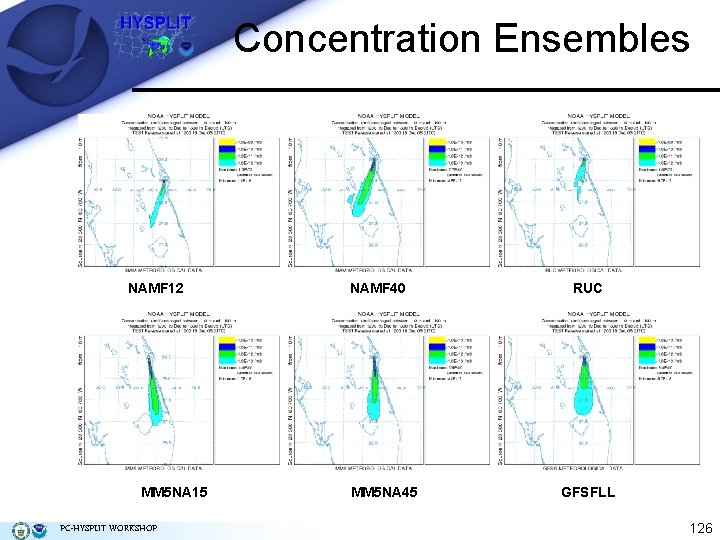
Concentration Ensembles NAMF 12 MM 5 NA 15 PC-HYSPLIT WORKSHOP NAMF 40 MM 5 NA 45 RUC GFSFLL 126

Concentration Ensembles Now, before running the ensemble display program the base name of the concentration files that were just created (cdump. 001 to cdump. 006) MUST be reset to the default in the Definition of Concentration Grid 1 menu. Change the last one selected, in this case cdump. 006 to cdump (this is automatically taken care of by the GUI if running a one meteorological dataset internal ensemble as will be shown later). Save the setup, but do not rerun the model. This creates a new CONTROL file with the proper base name cdump to be created by the ensemble program. Now, select View Map from the Concentration / Display Options / Ensemble menu. PC-HYSPLIT WORKSHOP 127

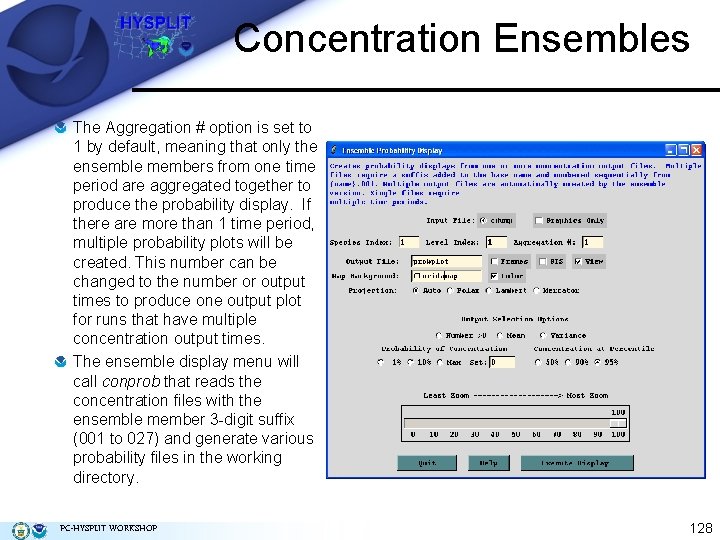
Concentration Ensembles The Aggregation # option is set to 1 by default, meaning that only the ensemble members from one time period are aggregated together to produce the probability display. If there are more than 1 time period, multiple probability plots will be created. This number can be changed to the number or output times to produce one output plot for runs that have multiple concentration output times. The ensemble display menu will call conprob that reads the concentration files with the ensemble member 3 -digit suffix (001 to 027) and generate various probability files in the working directory. PC-HYSPLIT WORKSHOP 128

Concentration Ensembles Definition of Outputs 1 - Number of ensemble members at each grid point with concentrations > 0. 2 - The Mean of all ensemble members. 3 – Variance (mean square difference between members and the mean). 4 – The Probability of Concentration produces contours that give the probability of exceeding a fixed concentration value at one of 3 levels: 1% of maximum, 10% of maximum and the maximum. The concentration level is displayed in the pollutant identification label like C 14, where 14 is the concentration to the power of 10 -14 5 – The Concentration at Percentile shows contours of areas where concentrations will be exceeded only at the given probability level (in the GUI these are 50, 90 and 95%). Click the Help button for details on each of these settings. PC-HYSPLIT WORKSHOP 129

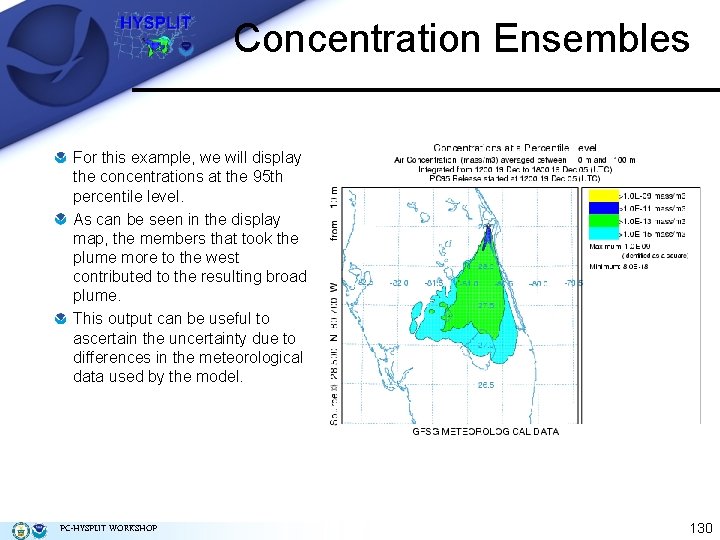
Concentration Ensembles For this example, we will display the concentrations at the 95 th percentile level. As can be seen in the display map, the members that took the plume more to the west contributed to the resulting broad plume. This output can be useful to ascertain the uncertainty due to differences in the meteorological data used by the model. PC-HYSPLIT WORKSHOP 130

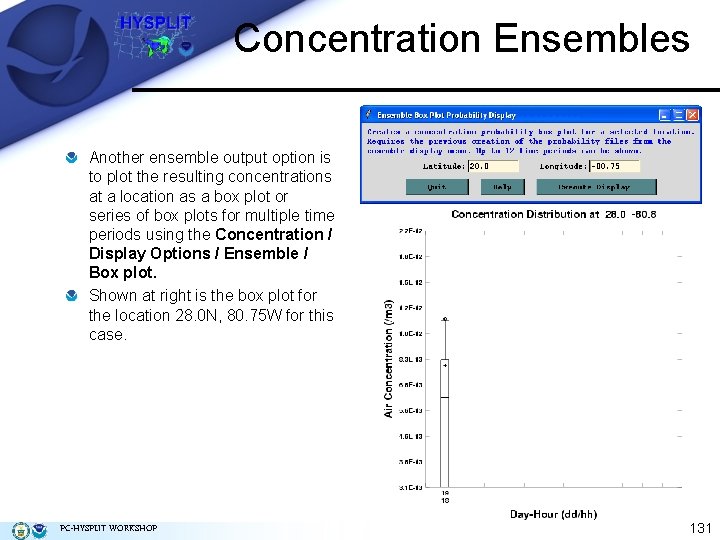
Concentration Ensembles Another ensemble output option is to plot the resulting concentrations at a location as a box plot or series of box plots for multiple time periods using the Concentration / Display Options / Ensemble / Box plot. Shown at right is the box plot for the location 28. 0 N, 80. 75 W for this case. PC-HYSPLIT WORKSHOP 131

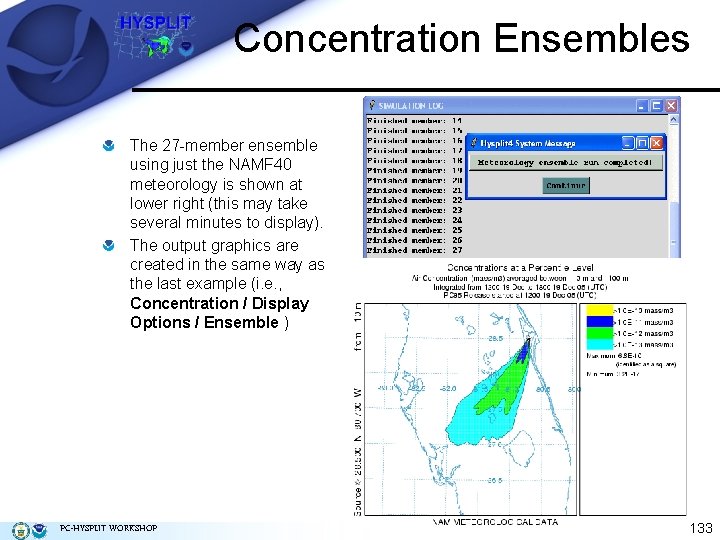
Concentration Ensembles Another approach to concentration ensembles is to generate an internal ensemble from a single meteorological data set. This computation is part of HYSPLIT and can be selected as the Ensemble option from the Concentration / Special Runs menu tab. In these simulations the meteorological data are perturbed to test the sensitivity of the simulation to the flow field. The meteorological grid is offset in either X, Y, or Z for each member of the ensemble. The calculation offset for each member of the ensemble is determined by the grid factor and can be adjusted in the Advanced / Configuration Setup / Concentration menu. The default offset is one meteorological grid point in the horizontal and 0. 01 sigma units in the vertical. The result is twenty-seven ensemble members for all offsets. Because the ensemble calculation offsets the starting point, it is suggested that for ground-level sources, the starting point height should be at least 0. 01 sigma (about 250 m) above the ground. PC-HYSPLIT WORKSHOP 132

Concentration Ensembles The 27 -member ensemble using just the NAMF 40 meteorology is shown at lower right (this may take several minutes to display). The output graphics are created in the same way as the last example (i. e. , Concentration / Display Options / Ensemble ) PC-HYSPLIT WORKSHOP 133

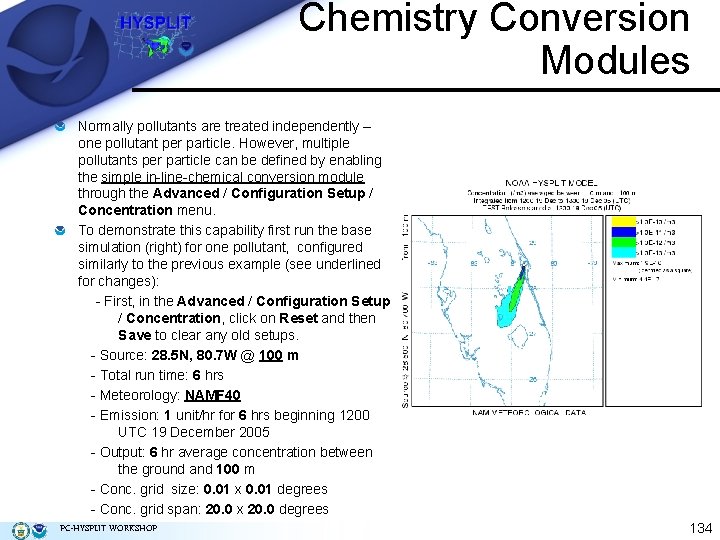
Chemistry Conversion Modules Normally pollutants are treated independently – one pollutant per particle. However, multiple pollutants per particle can be defined by enabling the simple in-line-chemical conversion module through the Advanced / Configuration Setup / Concentration menu. To demonstrate this capability first run the base simulation (right) for one pollutant, configured similarly to the previous example (see underlined for changes): - First, in the Advanced / Configuration Setup / Concentration, click on Reset and then Save to clear any old setups. - Source: 28. 5 N, 80. 7 W @ 100 m - Total run time: 6 hrs - Meteorology: NAMF 40 - Emission: 1 unit/hr for 6 hrs beginning 1200 UTC 19 December 2005 - Output: 6 hr average concentration between the ground and 100 m - Conc. grid size: 0. 01 x 0. 01 degrees - Conc. grid span: 20. 0 x 20. 0 degrees PC-HYSPLIT WORKSHOP 134

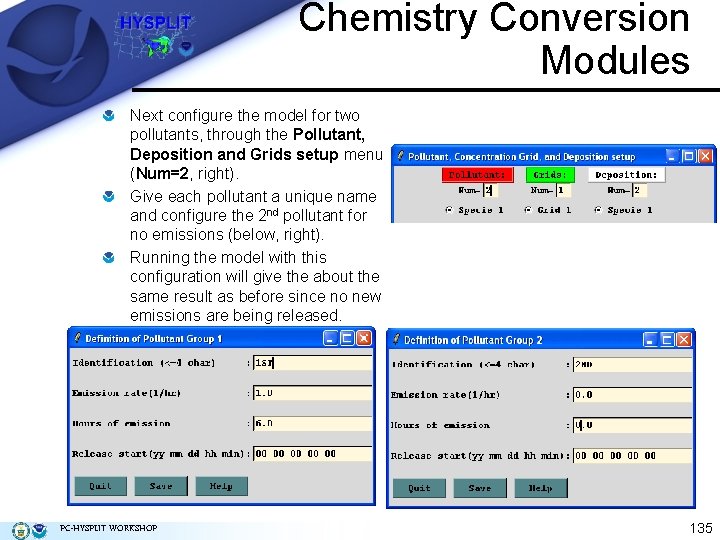
Chemistry Conversion Modules Next configure the model for two pollutants, through the Pollutant, Deposition and Grids setup menu (Num=2, right). Give each pollutant a unique name and configure the 2 nd pollutant for no emissions (below, right). Running the model with this configuration will give the about the same result as before since no new emissions are being released. PC-HYSPLIT WORKSHOP 135

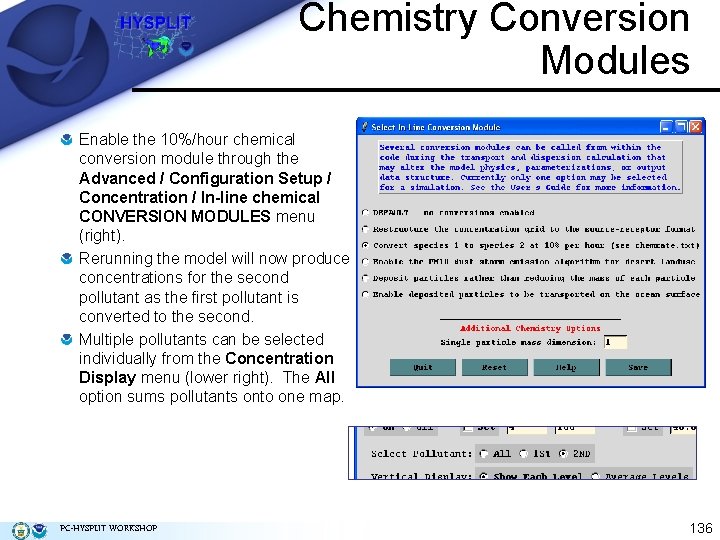
Chemistry Conversion Modules Enable the 10%/hour chemical conversion module through the Advanced / Configuration Setup / Concentration / In-line chemical CONVERSION MODULES menu (right). Rerunning the model will now produce concentrations for the second pollutant as the first pollutant is converted to the second. Multiple pollutants can be selected individually from the Concentration Display menu (lower right). The All option sums pollutants onto one map. PC-HYSPLIT WORKSHOP 136

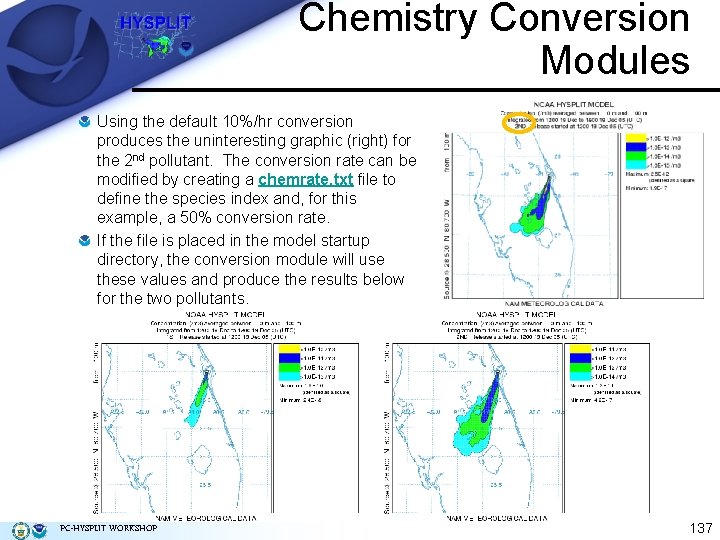
Chemistry Conversion Modules Using the default 10%/hr conversion produces the uninteresting graphic (right) for the 2 nd pollutant. The conversion rate can be modified by creating a chemrate. txt file to define the species index and, for this example, a 50% conversion rate. If the file is placed in the model startup directory, the conversion module will use these values and produce the results below for the two pollutants. PC-HYSPLIT WORKSHOP 137

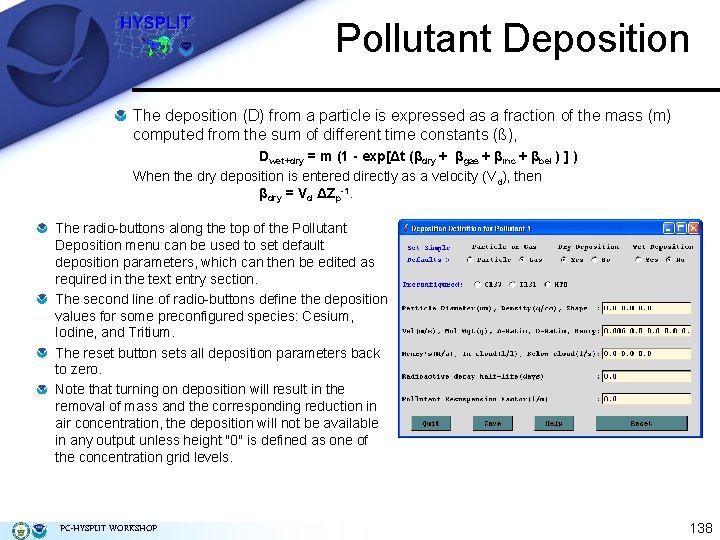
Pollutant Deposition The deposition (D) from a particle is expressed as a fraction of the mass (m) computed from the sum of different time constants (ß), Dwet+dry = m (1 - exp[Δt (βdry + βgas + βinc + βbel ) ] ) When the dry deposition is entered directly as a velocity (Vd), then βdry = Vd ΔZp-1. The radio-buttons along the top of the Pollutant Deposition menu can be used to set default deposition parameters, which can then be edited as required in the text entry section. The second line of radio-buttons define the deposition values for some preconfigured species: Cesium, Iodine, and Tritium. The reset button sets all deposition parameters back to zero. Note that turning on deposition will result in the removal of mass and the corresponding reduction in air concentration, the deposition will not be available in any output unless height "0" is defined as one of the concentration grid levels. PC-HYSPLIT WORKSHOP 138

Pollutant Deposition Dry Deposition: Dry deposition calculations are performed in the lowest model layer (75 m) based upon the relation that the deposition flux equals the velocity times the ground-level air concentration. This calculation is available for gases and particles. When dry deposition is entered directly as a velocity (Vd), then βdry = Vd ΔZp-1. Example: - First, click Reset and Save in the Advanced / Configuration Setup / Concentration menu to clear any old setups. - Source: 28. 5 N, 80. 7 W @ 10 m - Total run time: 24 hrs - Meteorology: NAMF 12 - Emission: 1 unit/hr for 1 hr beginning 1200 UTC 19 December 2005 - Output: 24 hr deposition (level 0) - Conc. grid size: 0. 05 x 0. 05 degrees - Conc. grid span: 20. 0 x 20. 0 degrees - Turn on dry deposition in the Deposition menu from the Pollutant, Deposition & Grids Setup menu (right). This automatically sets Vd to 0. 006 m/s for a gas. PC-HYSPLIT WORKSHOP 139

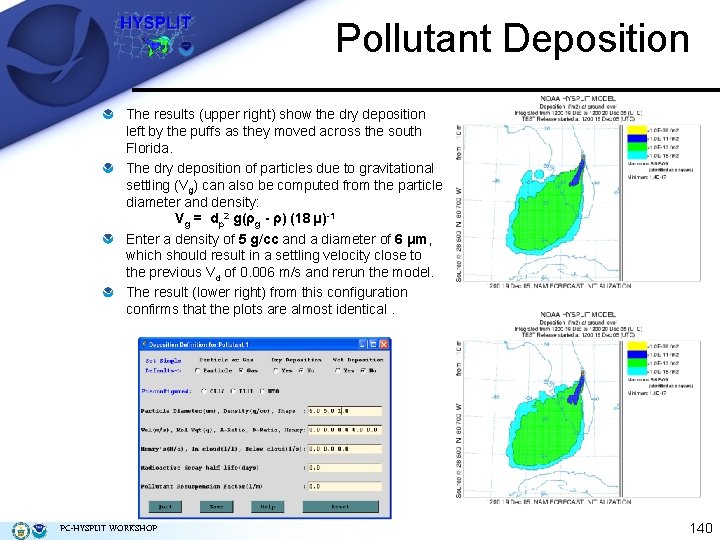
Pollutant Deposition The results (upper right) show the dry deposition left by the puffs as they moved across the south Florida. The dry deposition of particles due to gravitational settling (Vg) can also be computed from the particle diameter and density: Vg = dp 2 g(ρg - ρ) (18 µ)-1 Enter a density of 5 g/cc and a diameter of 6 μm, which should result in a settling velocity close to the previous Vd of 0. 006 m/s and rerun the model. The result (lower right) from this configuration confirms that the plots are almost identical. PC-HYSPLIT WORKSHOP 140

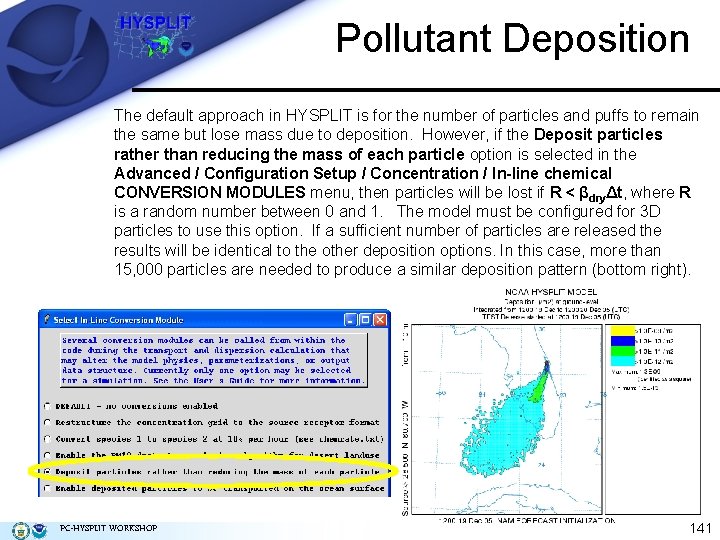
Pollutant Deposition The default approach in HYSPLIT is for the number of particles and puffs to remain the same but lose mass due to deposition. However, if the Deposit particles rather than reducing the mass of each particle option is selected in the Advanced / Configuration Setup / Concentration / In-line chemical CONVERSION MODULES menu, then particles will be lost if R < βdryΔt, where R is a random number between 0 and 1. The model must be configured for 3 D particles to use this option. If a sufficient number of particles are released the results will be identical to the other deposition options. In this case, more than 15, 000 particles are needed to produce a similar deposition pattern (bottom right). PC-HYSPLIT WORKSHOP 141

Pollutant Deposition Finally, the dry deposition velocity can also be calculated by the model using the resistance method, which requires setting the four parameters: molecular weight, surface reactivity, diffusivity, and the effective Henry's constant. See the table in the Help for more suggested numbers for some common pollutants. Radioactive decay can be specified by entering a value in days for the decay rate. A non-zero value in this field initiates the decay process of both airborne and deposited pollutants. A non-zero value for the re-suspension factor causes deposited pollutants to be re-emitted based upon soil conditions, wind velocity, and particle type. Pollutant re-suspension requires the definition of a deposition grid, as the pollutant is re-emitted from previously deposited material. Under most circumstances, the deposition should be accumulated on the grid for the entire duration of the simulation. Note that the air concentration and deposition grids may be defined at different temporal and spatial scales. PC-HYSPLIT WORKSHOP 142

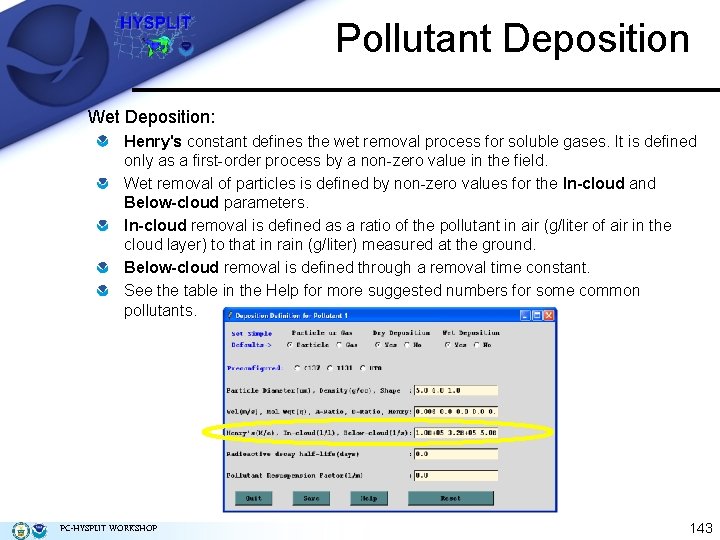
Pollutant Deposition Wet Deposition: Henry's constant defines the wet removal process for soluble gases. It is defined only as a first-order process by a non-zero value in the field. Wet removal of particles is defined by non-zero values for the In-cloud and Below-cloud parameters. In-cloud removal is defined as a ratio of the pollutant in air (g/liter of air in the cloud layer) to that in rain (g/liter) measured at the ground. Below-cloud removal is defined through a removal time constant. See the table in the Help for more suggested numbers for some common pollutants. PC-HYSPLIT WORKSHOP 143

Source Attribution using Back Trajectory Analysis Frequently it is necessary to attribute a pollutant measurement to a specific source location. One approach is to compute a backward trajectory to determine the air’s origin. Although it is not uncommon to see sources identified by a single trajectory, the uncertainties inherent in a single-trajectory can preempt its utility. One way to reduce those uncertainties would be to compute multiple trajectories, in height, time, and space. Case Study: High ozone event in Atlanta, Georgia on August 15, 2007. Daily Maximum 1 -hour ozone values of 139 ppb. PC-HYSPLIT WORKSHOP 144

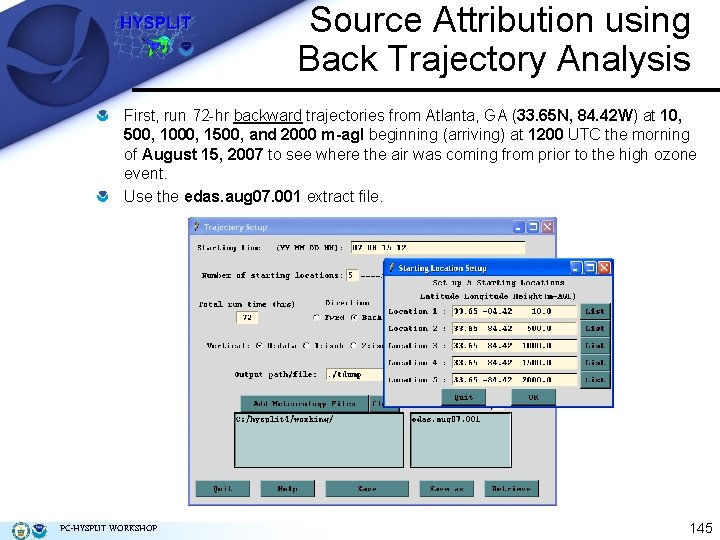
Source Attribution using Back Trajectory Analysis First, run 72 -hr backward trajectories from Atlanta, GA (33. 65 N, 84. 42 W) at 10, 500, 1000, 1500, and 2000 m-agl beginning (arriving) at 1200 UTC the morning of August 15, 2007 to see where the air was coming from prior to the high ozone event. Use the edas. aug 07. 001 extract file. PC-HYSPLIT WORKSHOP 145

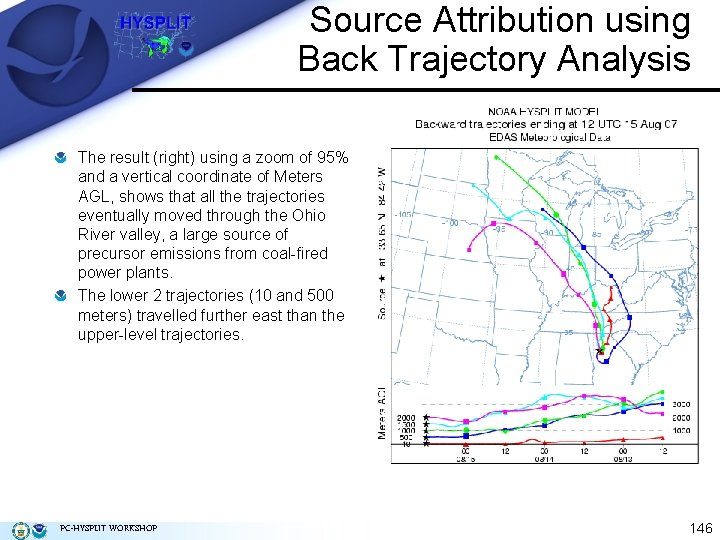
Source Attribution using Back Trajectory Analysis The result (right) using a zoom of 95% and a vertical coordinate of Meters AGL, shows that all the trajectories eventually moved through the Ohio River valley, a large source of precursor emissions from coal-fired power plants. The lower 2 trajectories (10 and 500 meters) travelled further east than the upper-level trajectories. PC-HYSPLIT WORKSHOP 146

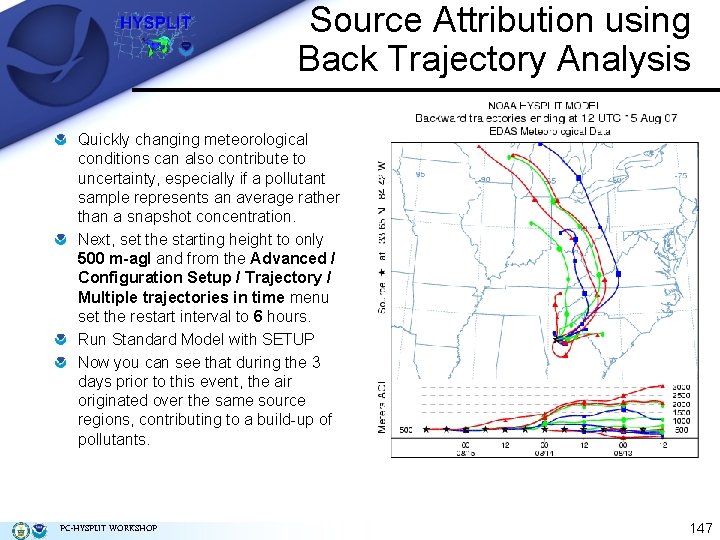
Source Attribution using Back Trajectory Analysis Quickly changing meteorological conditions can also contribute to uncertainty, especially if a pollutant sample represents an average rather than a snapshot concentration. Next, set the starting height to only 500 m-agl and from the Advanced / Configuration Setup / Trajectory / Multiple trajectories in time menu set the restart interval to 6 hours. Run Standard Model with SETUP Now you can see that during the 3 days prior to this event, the air originated over the same source regions, contributing to a build-up of pollutants. PC-HYSPLIT WORKSHOP 147

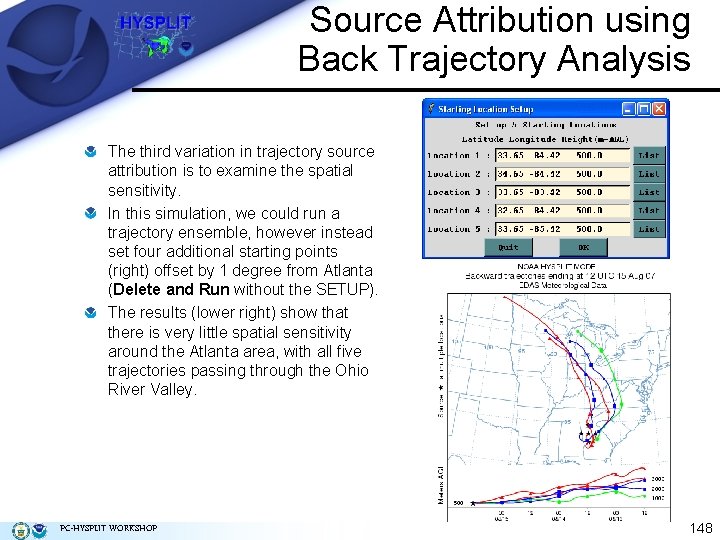
Source Attribution using Back Trajectory Analysis The third variation in trajectory source attribution is to examine the spatial sensitivity. In this simulation, we could run a trajectory ensemble, however instead set four additional starting points (right) offset by 1 degree from Atlanta (Delete and Run without the SETUP). The results (lower right) show that there is very little spatial sensitivity around the Atlanta area, with all five trajectories passing through the Ohio River Valley. PC-HYSPLIT WORKSHOP 148

Source Attribution using Source-Receptor Matrices The term “matrix” has two connotations with respect to HYSPLIT. In the earlier application, a matrix of sources was created, the results of which were summed to a single concentration file. In this application, defining a concentration grid for each source creates a matrix of sources and receptors. For this simulation, we are interested in knowing what fraction of the 6 -hourly average air concentration at Atlanta was contributed by a grid of source locations within the Ohio Valley region, assuming that there are no other contributions from other sources. First we will lay out a grid of source locations over the Ohio Valley between 35 N, 95 W and 45 N, 75 W at 1 degree intervals. Then we will release 500 3 D particles from each source location over the 72 hour simulation at 50 m-agl and determine the contribution to Atlanta from these sources. PC-HYSPLIT WORKSHOP 149

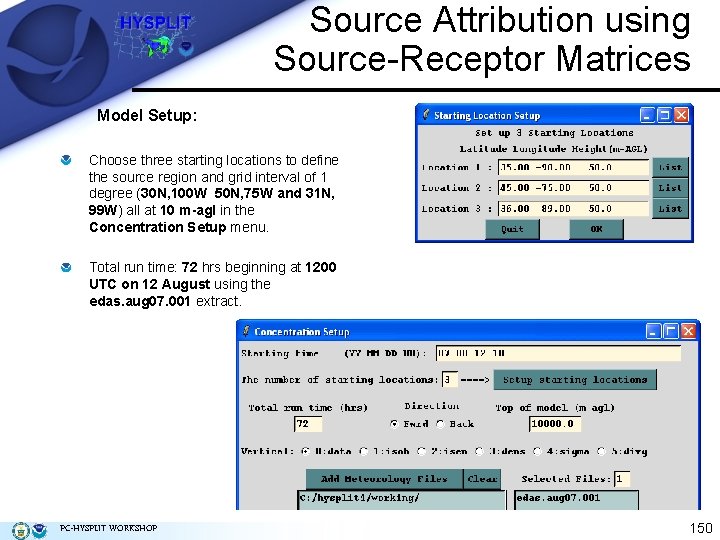
Source Attribution using Source-Receptor Matrices Model Setup: Choose three starting locations to define the source region and grid interval of 1 degree (30 N, 100 W 50 N, 75 W and 31 N, 99 W) all at 10 m-agl in the Concentration Setup menu. Total run time: 72 hrs beginning at 1200 UTC on 12 August using the edas. aug 07. 001 extract. PC-HYSPLIT WORKSHOP 150

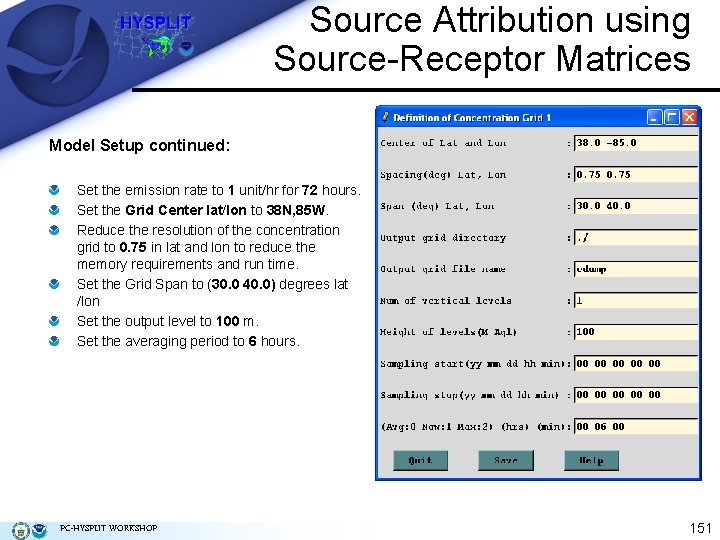
Source Attribution using Source-Receptor Matrices Model Setup continued: Set the emission rate to 1 unit/hr for 72 hours. Set the Grid Center lat/lon to 38 N, 85 W. Reduce the resolution of the concentration grid to 0. 75 in lat and lon to reduce the memory requirements and run time. Set the Grid Span to (30. 0 40. 0) degrees lat /lon Set the output level to 100 m. Set the averaging period to 6 hours. PC-HYSPLIT WORKSHOP 151

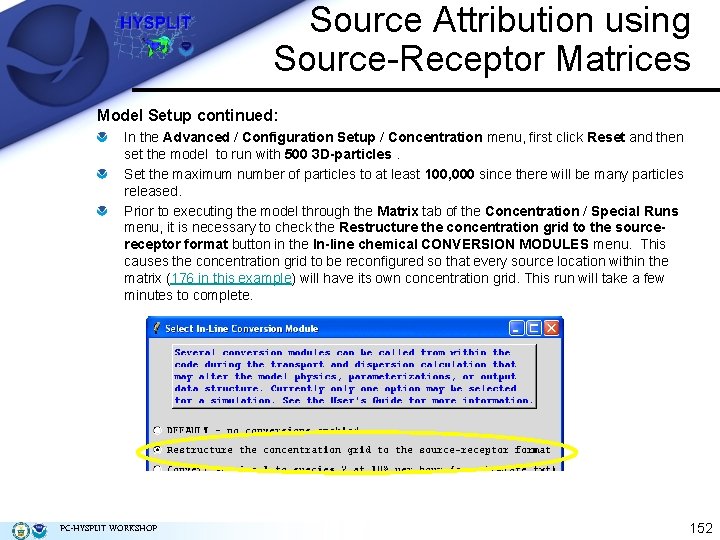
Source Attribution using Source-Receptor Matrices Model Setup continued: In the Advanced / Configuration Setup / Concentration menu, first click Reset and then set the model to run with 500 3 D-particles. Set the maximum number of particles to at least 100, 000 since there will be many particles released. Prior to executing the model through the Matrix tab of the Concentration / Special Runs menu, it is necessary to check the Restructure the concentration grid to the sourcereceptor format button in the In-line chemical CONVERSION MODULES menu. This causes the concentration grid to be reconfigured so that every source location within the matrix (176 in this example) will have its own concentration grid. This run will take a few minutes to complete. PC-HYSPLIT WORKSHOP 152

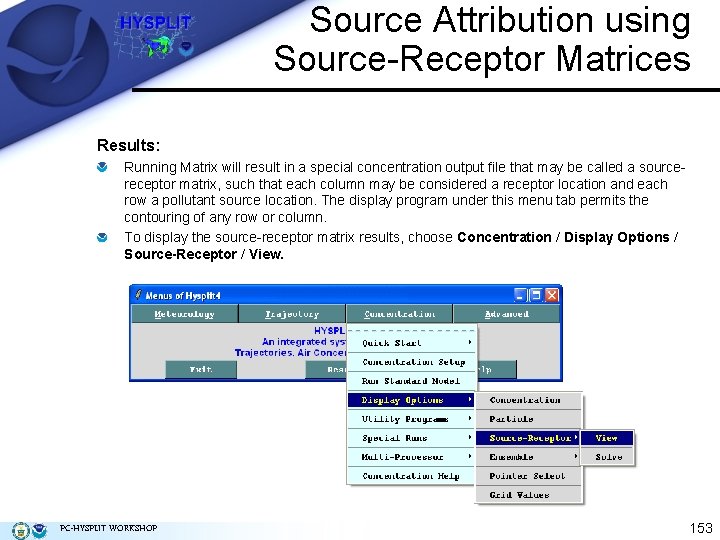
Source Attribution using Source-Receptor Matrices Results: Running Matrix will result in a special concentration output file that may be called a sourcereceptor matrix, such that each column may be considered a receptor location and each row a pollutant source location. The display program under this menu tab permits the contouring of any row or column. To display the source-receptor matrix results, choose Concentration / Display Options / Source-Receptor / View. PC-HYSPLIT WORKSHOP 153

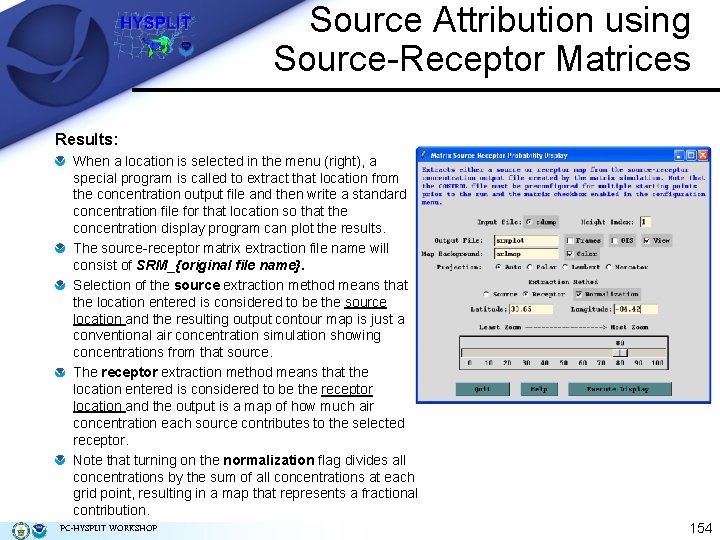
Source Attribution using Source-Receptor Matrices Results: When a location is selected in the menu (right), a special program is called to extract that location from the concentration output file and then write a standard concentration file for that location so that the concentration display program can plot the results. The source-receptor matrix extraction file name will consist of SRM_{original file name}. Selection of the source extraction method means that the location entered is considered to be the source location and the resulting output contour map is just a conventional air concentration simulation showing concentrations from that source. The receptor extraction method means that the location entered is considered to be the receptor location and the output is a map of how much air concentration each source contributes to the selected receptor. Note that turning on the normalization flag divides all concentrations by the sum of all concentrations at each grid point, resulting in a map that represents a fractional contribution. PC-HYSPLIT WORKSHOP 154

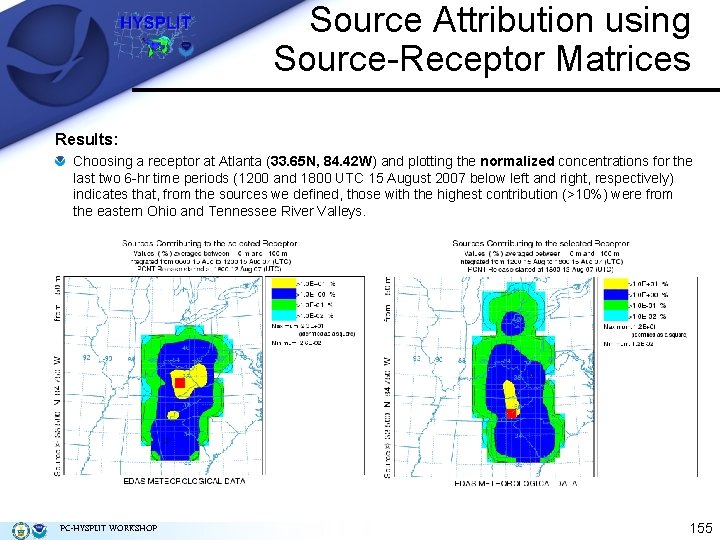
Source Attribution using Source-Receptor Matrices Results: Choosing a receptor at Atlanta (33. 65 N, 84. 42 W) and plotting the normalized concentrations for the last two 6 -hr time periods (1200 and 1800 UTC 15 August 2007 below left and right, respectively) indicates that, from the sources we defined, those with the highest contribution (>10%) were from the eastern Ohio and Tennessee River Valleys. PC-HYSPLIT WORKSHOP 155

Source Attribution Functions Running the air concentration prediction model backwards is comparable to a back trajectory calculation but it includes the dispersion component of the calculation. The result, although it looks like an air concentration field, is more comparable to a source attribution function. If the atmospheric turbulence were stationary and homogeneous then this attribution function would yield the same result from receptor to source as a forward calculation from source to receptor. PC-HYSPLIT WORKSHOP 156

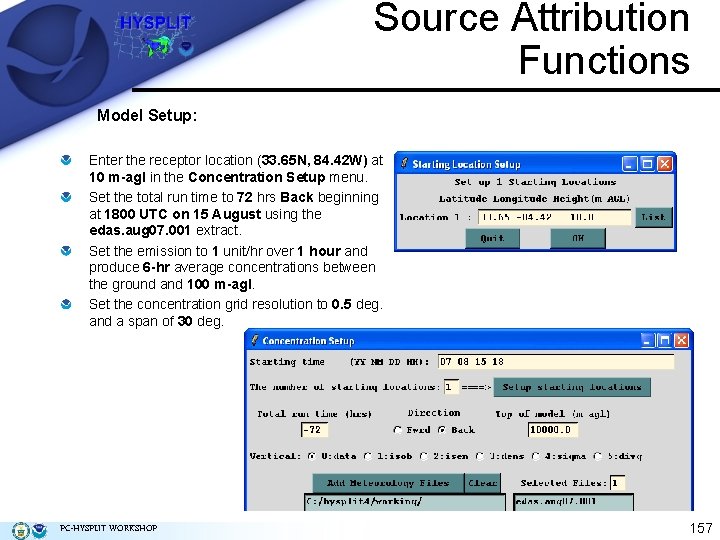
Source Attribution Functions Model Setup: Enter the receptor location (33. 65 N, 84. 42 W) at 10 m-agl in the Concentration Setup menu. Set the total run time to 72 hrs Back beginning at 1800 UTC on 15 August using the edas. aug 07. 001 extract. Set the emission to 1 unit/hr over 1 hour and produce 6 -hr average concentrations between the ground and 100 m-agl. Set the concentration grid resolution to 0. 5 deg. and a span of 30 deg. PC-HYSPLIT WORKSHOP 157

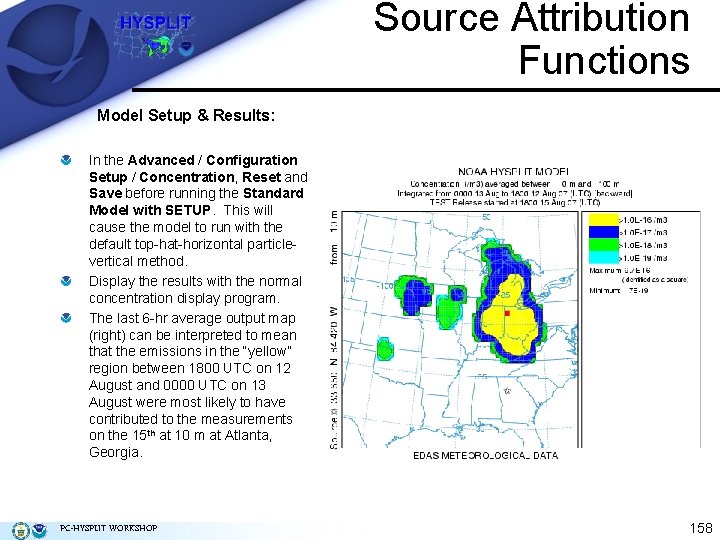
Source Attribution Functions Model Setup & Results: In the Advanced / Configuration Setup / Concentration, Reset and Save before running the Standard Model with SETUP. This will cause the model to run with the default top-hat-horizontal particlevertical method. Display the results with the normal concentration display program. The last 6 -hr average output map (right) can be interpreted to mean that the emissions in the “yellow” region between 1800 UTC on 12 August and 0000 UTC on 13 August were most likely to have contributed to the measurements on the 15 th at 10 m at Atlanta, Georgia. PC-HYSPLIT WORKSHOP 158

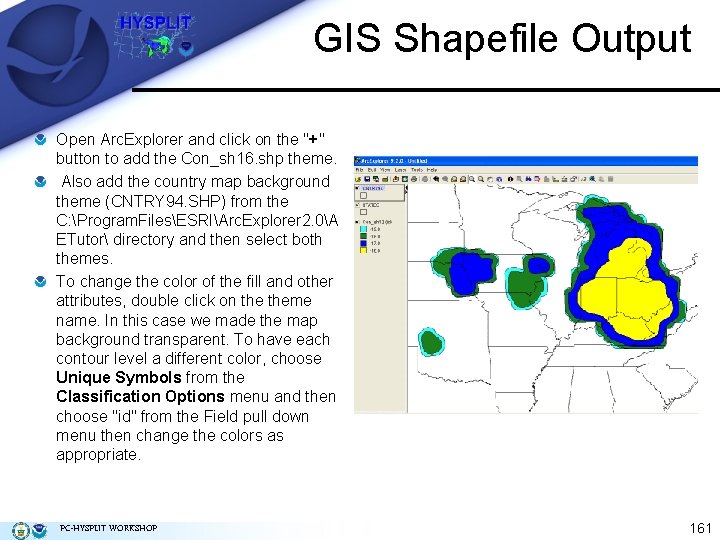
GIS Shapefile Output Graphical output from most GUI programs can be converted into an ESRI GIS shapefile format for use in GIS programs such as Arc. Explorer, Arc. GIS Explorer, and Google Earth. To convert the graphic generated in the last example, check the Frames and ESRI Generate boxes in the Concentration Display menu (below). This will result in a unique Postscript file and 2 text files for each time period (ignore the concplot. ps not found message). The Generate format text output file (GIS_00100_ps_HH. txt, where HH is the sequence number) contains the latitude and longitude pairs that make up each contour. From Concentration / Utility Programs, use the GIS to Shapefile menu to select text files. They are named by level and time sequence. Select the last file (GIS_00100_ps_12. txt) and rename the output file to reflect the same number (Con_sh 12). Make sure the Conversion method is set to polygons, since we are outputting concentration contours. When finished there will be a series of files with the suffix “shp”, “shx”, and “dbf” in the working directory. PC-HYSPLIT WORKSHOP 159

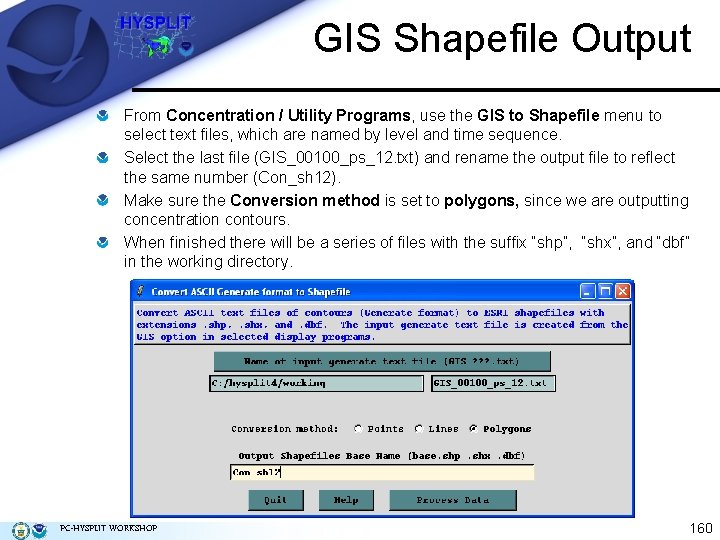
GIS Shapefile Output From Concentration / Utility Programs, use the GIS to Shapefile menu to select text files, which are named by level and time sequence. Select the last file (GIS_00100_ps_12. txt) and rename the output file to reflect the same number (Con_sh 12). Make sure the Conversion method is set to polygons, since we are outputting concentration contours. When finished there will be a series of files with the suffix “shp”, “shx”, and “dbf” in the working directory. PC-HYSPLIT WORKSHOP 160

GIS Shapefile Output Open Arc. Explorer and click on the "+" button to add the Con_sh 16. shp theme. Also add the country map background theme (CNTRY 94. SHP) from the C: Program. FilesESRIArc. Explorer 2. 0A ETutor directory and then select both themes. To change the color of the fill and other attributes, double click on theme name. In this case we made the map background transparent. To have each contour level a different color, choose Unique Symbols from the Classification Options menu and then choose "id" from the Field pull down menu then change the colors as appropriate. PC-HYSPLIT WORKSHOP 161

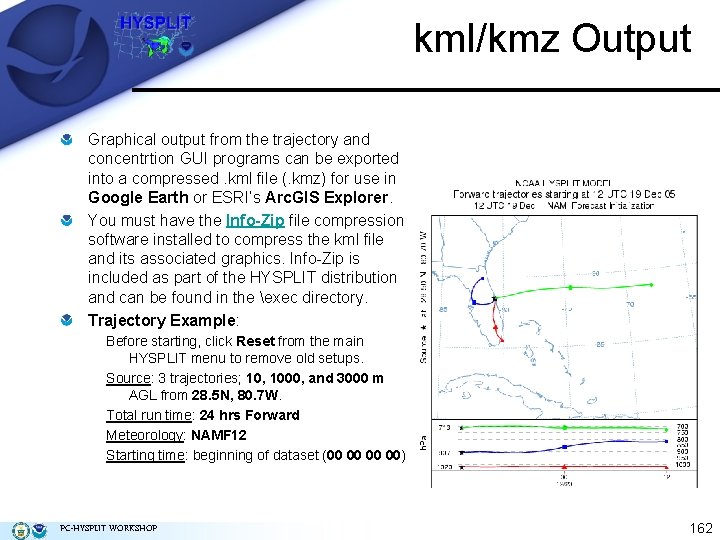
kml/kmz Output Graphical output from the trajectory and concentrtion GUI programs can be exported into a compressed. kml file (. kmz) for use in Google Earth or ESRI’s Arc. GIS Explorer. You must have the Info-Zip file compression software installed to compress the kml file and its associated graphics. Info-Zip is included as part of the HYSPLIT distribution and can be found in the exec directory. Trajectory Example: Before starting, click Reset from the main HYSPLIT menu to remove old setups. Source: 3 trajectories; 10, 1000, and 3000 m AGL from 28. 5 N, 80. 7 W. Total run time: 24 hrs Forward Meteorology: NAMF 12 Starting time: beginning of dataset (00 00) PC-HYSPLIT WORKSHOP 162

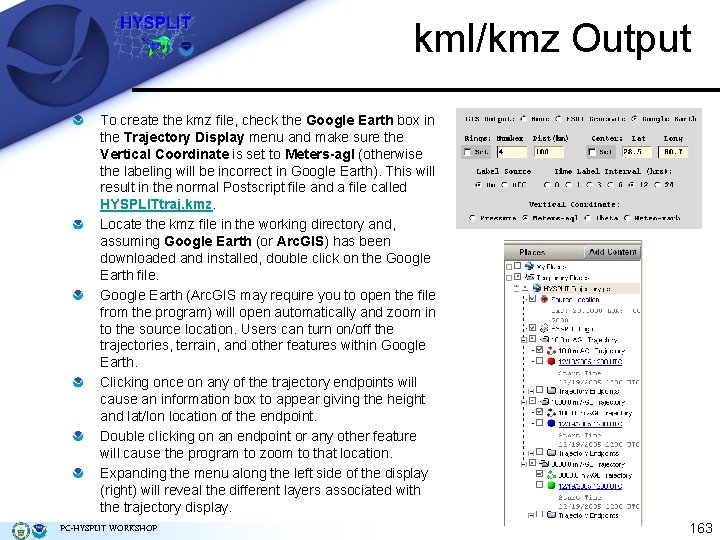
kml/kmz Output To create the kmz file, check the Google Earth box in the Trajectory Display menu and make sure the Vertical Coordinate is set to Meters-agl (otherwise the labeling will be incorrect in Google Earth). This will result in the normal Postscript file and a file called HYSPLITtraj. kmz. Locate the kmz file in the working directory and, assuming Google Earth (or Arc. GIS) has been downloaded and installed, double click on the Google Earth file. Google Earth (Arc. GIS may require you to open the file from the program) will open automatically and zoom in to the source location. Users can turn on/off the trajectories, terrain, and other features within Google Earth. Clicking once on any of the trajectory endpoints will cause an information box to appear giving the height and lat/lon location of the endpoint. Double clicking on an endpoint or any other feature will cause the program to zoom to that location. Expanding the menu along the left side of the display (right) will reveal the different layers associated with the trajectory display. PC-HYSPLIT WORKSHOP 163

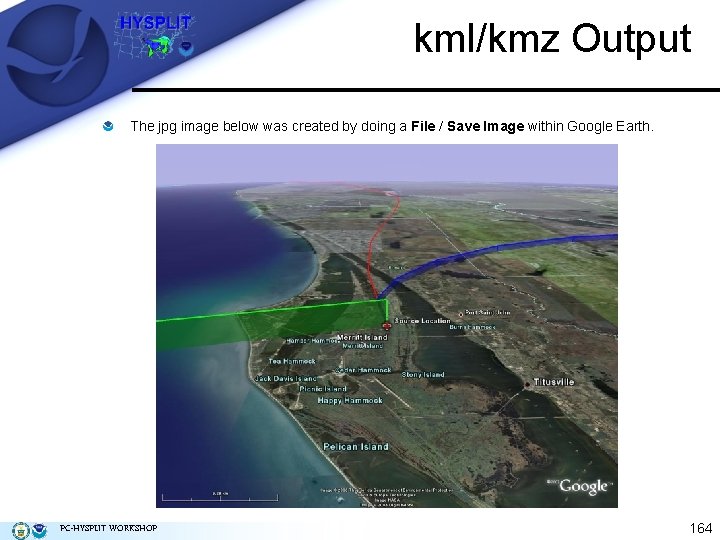
kml/kmz Output The jpg image below was created by doing a File / Save Image within Google Earth. PC-HYSPLIT WORKSHOP 164

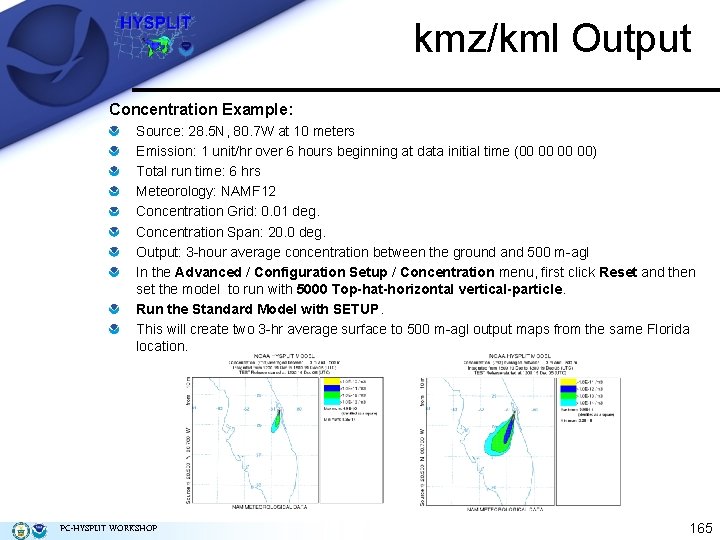
kmz/kml Output Concentration Example: Source: 28. 5 N, 80. 7 W at 10 meters Emission: 1 unit/hr over 6 hours beginning at data initial time (00 00) Total run time: 6 hrs Meteorology: NAMF 12 Concentration Grid: 0. 01 deg. Concentration Span: 20. 0 deg. Output: 3 -hour average concentration between the ground and 500 m-agl In the Advanced / Configuration Setup / Concentration menu, first click Reset and then set the model to run with 5000 Top-hat-horizontal vertical-particle. Run the Standard Model with SETUP. This will create two 3 -hr average surface to 500 m-agl output maps from the same Florida location. PC-HYSPLIT WORKSHOP 165

kmz/kml Output Concentration Example: To create the Google Earth formatted file check the Google Earth box in the Concentration Display menu. This will result in the normal Postscript file and a file called HYSPLITconc. kmz. Opening the Google Earth file results in 2 plumes to display (0 -3 h and 3 -6 h averages). Animate the image by using the VCR buttons at the top of the display The image below shows the 3 -6 h average between the ground and 500 m AGL. Using the controls in Google Earth allows the user to rotate, pan, and zoom the plume. Click on the second 3 h average plume link on the left menu to overlay it on the 0 -3 h average plume. PC-HYSPLIT WORKSHOP 166

kmz/kml Output Finally, as an example to show the 3 -dimensional terrain (NOTE: the Google Earth terrain is different from the meteorological model terrain so that the model contours and trajectories may be below or above the shown terrain), a trajectory and a concentration run was produced from a location in the Grand Canyon. The concentration CONTROL and SETUP. CFG files can be used to reproduce the concentration Google Earth file, and the trajectory CONTROL and SETUP. CFG files can be used to reproduce the trajectory Google Earth file. PC-HYSPLIT WORKSHOP 167

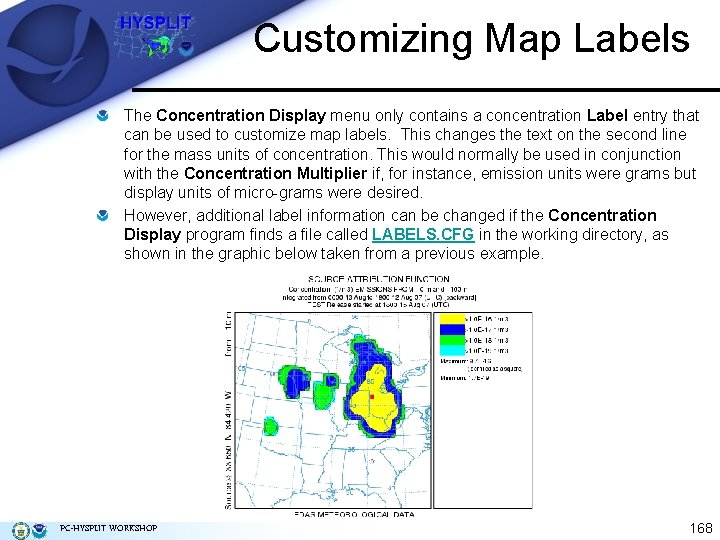
Customizing Map Labels The Concentration Display menu only contains a concentration Label entry that can be used to customize map labels. This changes the text on the second line for the mass units of concentration. This would normally be used in conjunction with the Concentration Multiplier if, for instance, emission units were grams but display units of micro-grams were desired. However, additional label information can be changed if the Concentration Display program finds a file called LABELS. CFG in the working directory, as shown in the graphic below taken from a previous example. PC-HYSPLIT WORKSHOP 168

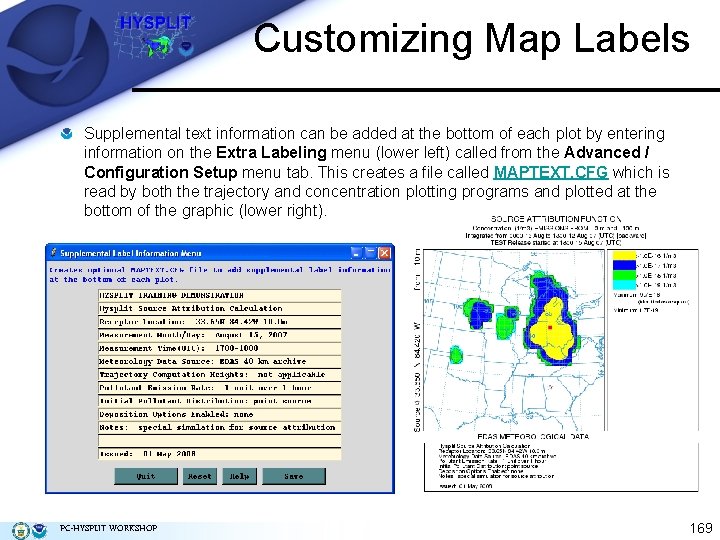
Customizing Map Labels Supplemental text information can be added at the bottom of each plot by entering information on the Extra Labeling menu (lower left) called from the Advanced / Configuration Setup menu tab. This creates a file called MAPTEXT. CFG which is read by both the trajectory and concentration plotting programs and plotted at the bottom of the graphic (lower right). PC-HYSPLIT WORKSHOP 169

Scripting for Automated Operations The guicode and examplesscripts directories contains example scripts that can be used to automate computations (auto_traj. tcl, auto_conc. tcl, auto_geol. tcl). Familiarity with the command line options is essential in modifying and writing new scripts in a text editor. Script syntax is very similar to the “C” language. All scripts work in the same manner by writing a new CONTROL file for each simulation, running the model, and then renaming the output. In this EXAMPLE SCRIPT, trajectories are computed at four locations and each is run separately for 24 hours using the NAMF 40 meteorological data. Place this script in the /working directory and double click on it to run it. Close the window when it appears and the trajectory Postscript files should be in the /working directory. PC-HYSPLIT WORKSHOP 170