Widespread parallel evolution Orthology Review of plant David










- Slides: 41

Widespread parallel evolution Orthology Review of plant David Emms david. emms@plants. ox. ac. uk Dept. Plant Sciences, University of Oxford Lausanne Nov 1 st 2019 https: //github. com/davidemms

All Species are Related by Common Descent • Any group of organisms share a common ancestor • Darwin, On the Origin of Species: Therefore I should infer from analogy that probably all the organic beings which have ever lived on this earth have descended from some one primordial form, into which life was first breathed.

Common Descent Implies Common Genes • Common descent implies common genes: • Species inherit genes from their ancestors • They must share genes in common • “Orthologs are genes in different species that can be traced to the same gene in their last common ancestral genome” (Glover et al 2019)

Orthologs Have Many Uses: Functional Annotation • Much research is carried out in model species • Mouse – model for human disease • Drosophila melanogaster – genetics • C. elegans – genetic control of development • Arabidopsis thaliana – model plant • Saccharomyces cerevisiae – single cell eukaryote • Approximately 0% of genes will be studied experimentally • Functions of unstudied genes often inferred from functions of orthologs • Identifying candidate genes Drosophila: André Karwath CC BY-SA 2. 5 S. cerevisiae: M. Das Murtey and P. Ramasamy CC BY-SA 3. 0

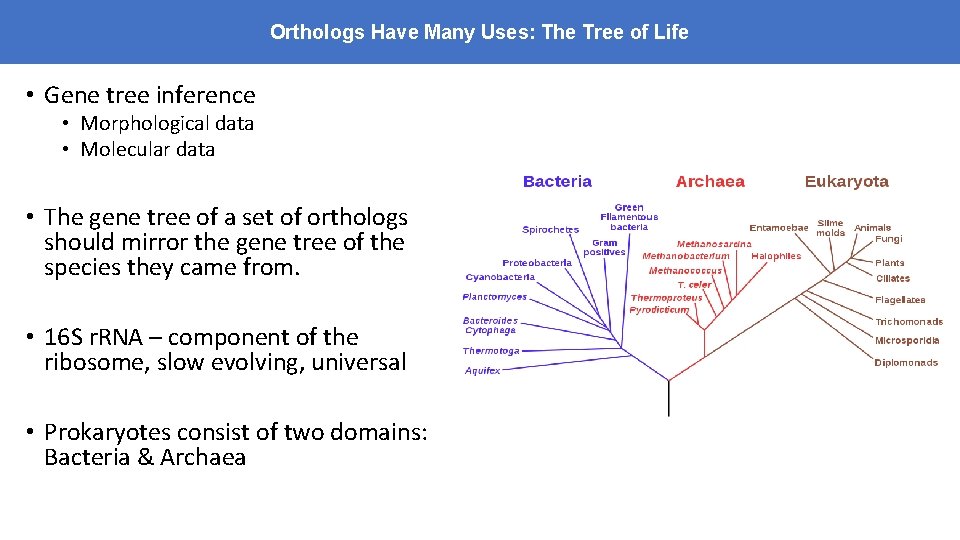
Orthologs Have Many Uses: The Tree of Life • Gene tree inference • Morphological data • Molecular data • The gene tree of a set of orthologs should mirror the gene tree of the species they came from. • 16 S r. RNA – component of the ribosome, slow evolving, universal • Prokaryotes consist of two domains: Bacteria & Archaea

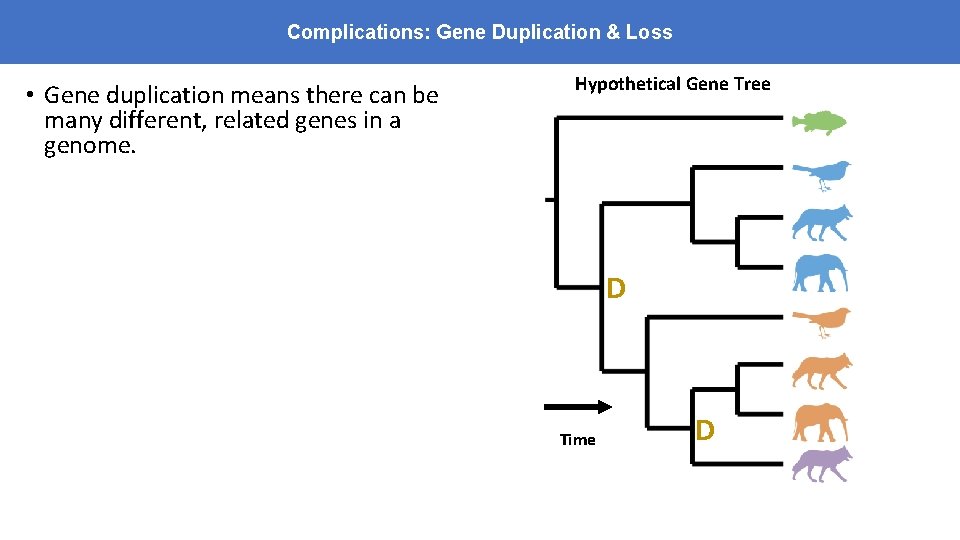
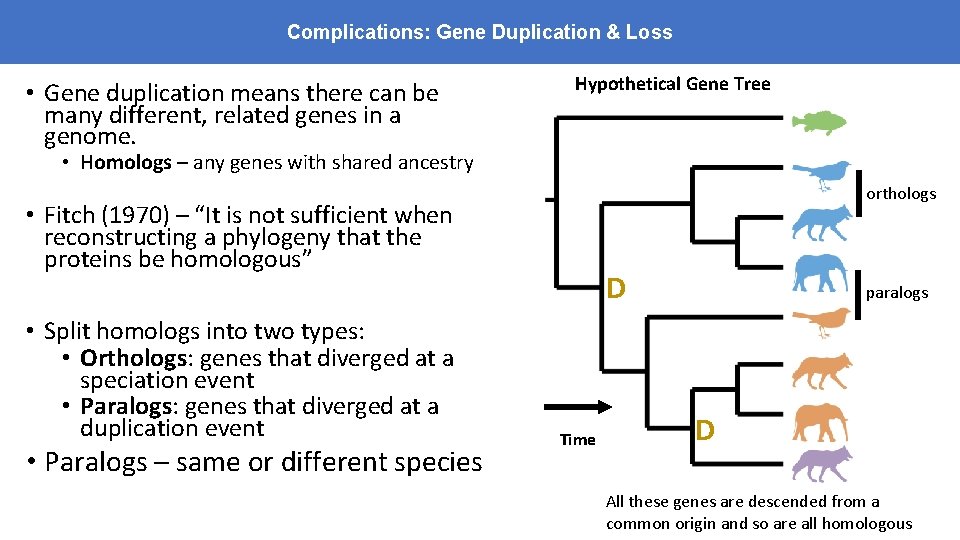
Complications: Gene Duplication & Loss • Gene duplication means there can be many different, related genes in a genome. Hypothetical Gene Tree • Homologs – any genes with shared ancestry • Fitch (1970) – “It is not sufficient when reconstructing a phylogeny that the proteins be homologous” • Split homologs into two types: • Orthologs: genes that diverged at a speciation event • Paralogs: genes that diverged at a duplication event D Time D

Complications: Gene Duplication & Loss • Gene duplication means there can be many different, related genes in a genome. Hypothetical Gene Tree • Homologs – any genes with shared ancestry orthologs • Fitch (1970) – “It is not sufficient when reconstructing a phylogeny that the proteins be homologous” • Split homologs into two types: • Orthologs: genes that diverged at a speciation event • Paralogs: genes that diverged at a duplication event • Paralogs – same or different species D Time paralogs D All these genes are descended from a common origin and so are all homologous

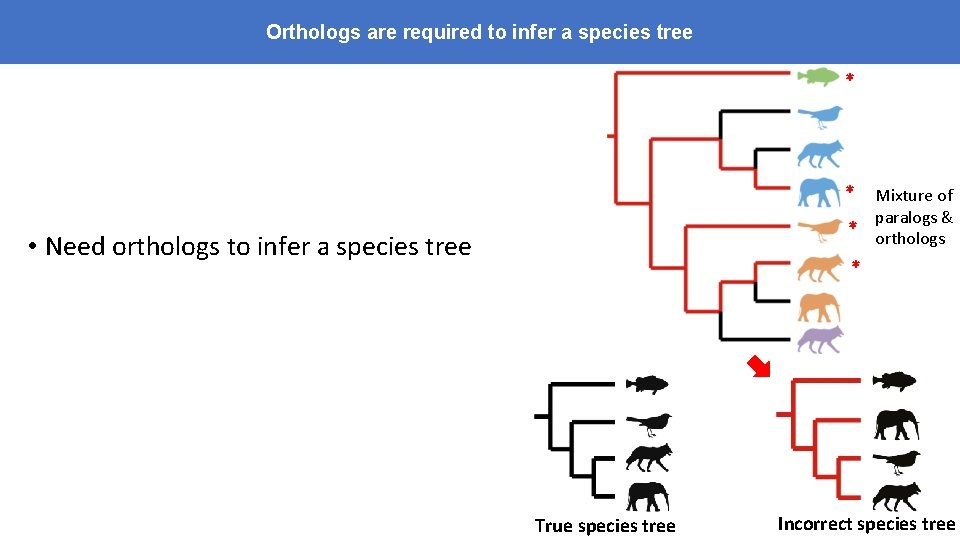
Orthologs are required to infer a species tree * * Mixture of paralogs & * orthologs • Need orthologs to infer a species tree * True species tree Incorrect species tree

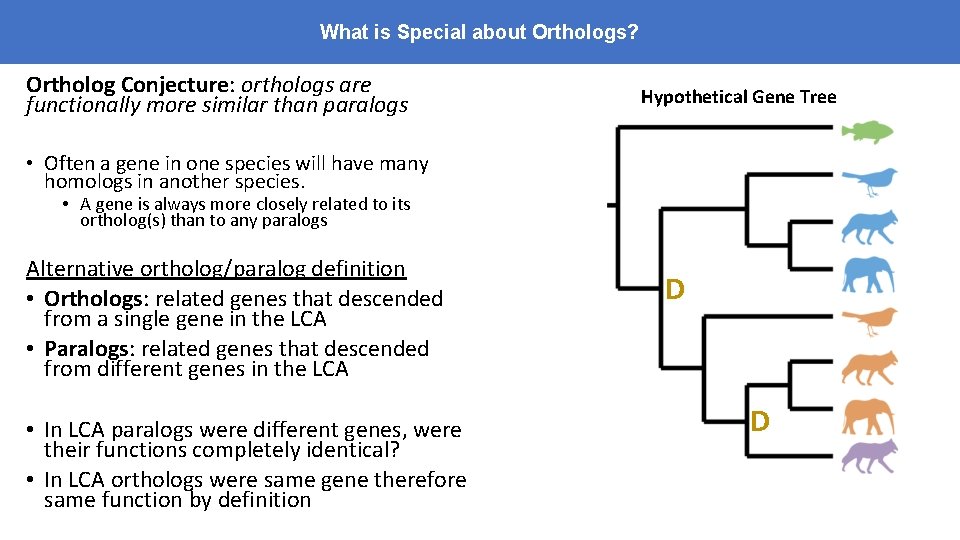
What is Special about Orthologs? Ortholog Conjecture: orthologs are functionally more similar than paralogs Hypothetical Gene Tree • Often a gene in one species will have many homologs in another species. • A gene is always more closely related to its ortholog(s) than to any paralogs Alternative ortholog/paralog definition • Orthologs: related genes that descended from a single gene in the LCA • Paralogs: related genes that descended from different genes in the LCA • In LCA paralogs were different genes, were their functions completely identical? • In LCA orthologs were same gene therefore same function by definition D D

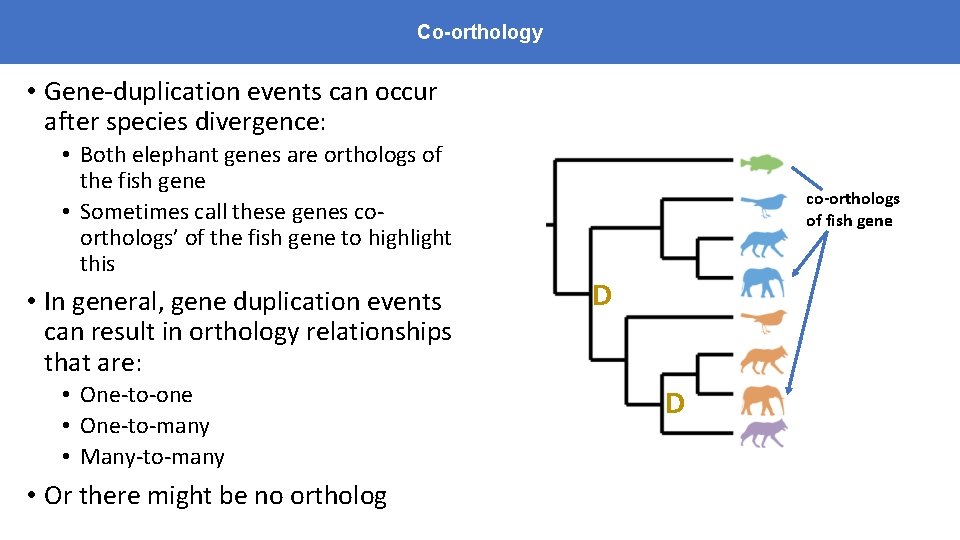
Co-orthology • Gene-duplication events can occur after species divergence: • Both elephant genes are orthologs of the fish gene • Sometimes call these genes coorthologs’ of the fish gene to highlight this • In general, gene duplication events can result in orthology relationships that are: • One-to-one • One-to-many • Many-to-many • Or there might be no ortholog co-orthologs of fish gene D D

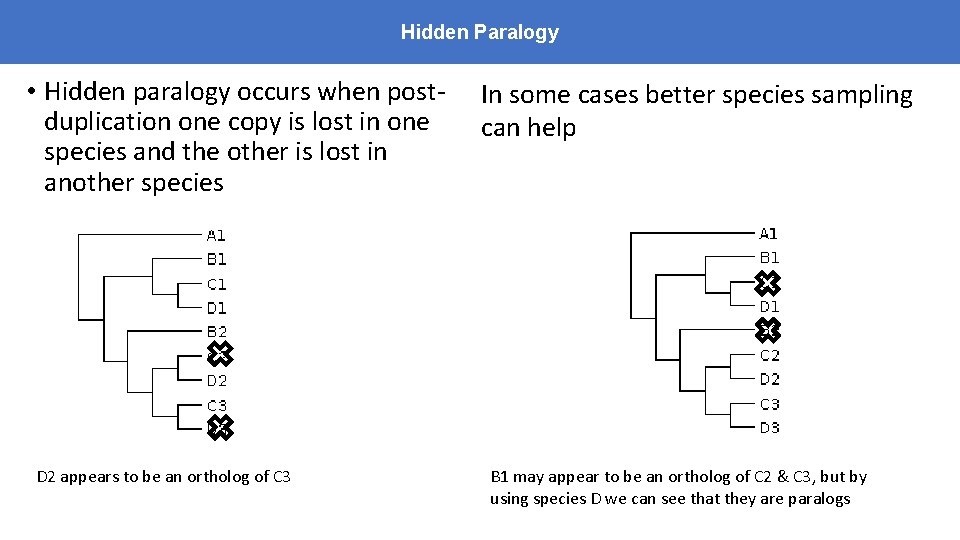
Hidden Paralogy • Hidden paralogy occurs when postduplication one copy is lost in one species and the other is lost in another species D 2 appears to be an ortholog of C 3 In some cases better species sampling can help B 1 may appear to be an ortholog of C 2 & C 3, but by using species D we can see that they are paralogs

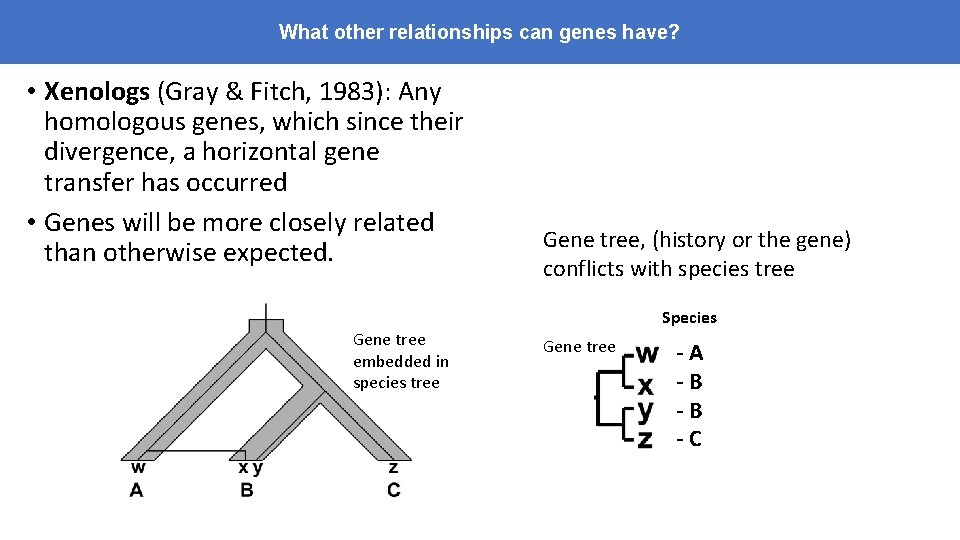
What other relationships can genes have? • Xenologs (Gray & Fitch, 1983): Any homologous genes, which since their divergence, a horizontal gene transfer has occurred • Genes will be more closely related than otherwise expected. Gene tree, (history or the gene) conflicts with species tree Species Gene tree embedded in species tree Gene tree -A -B -B -C

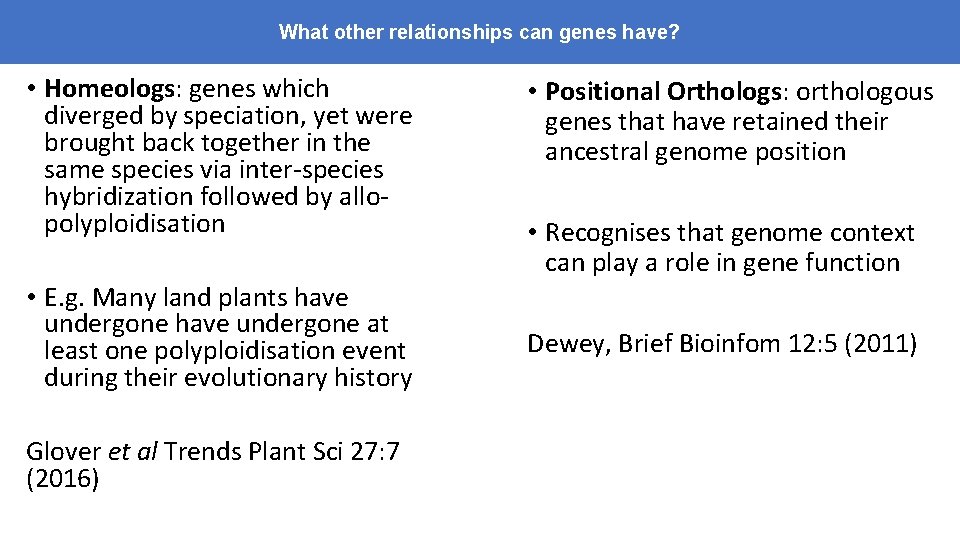
What other relationships can genes have? • Homeologs: genes which diverged by speciation, yet were brought back together in the same species via inter-species hybridization followed by allopolyploidisation • E. g. Many land plants have undergone at least one polyploidisation event during their evolutionary history Glover et al Trends Plant Sci 27: 7 (2016) • Positional Orthologs: orthologous genes that have retained their ancestral genome position • Recognises that genome context can play a role in gene function Dewey, Brief Bioinfom 12: 5 (2011)

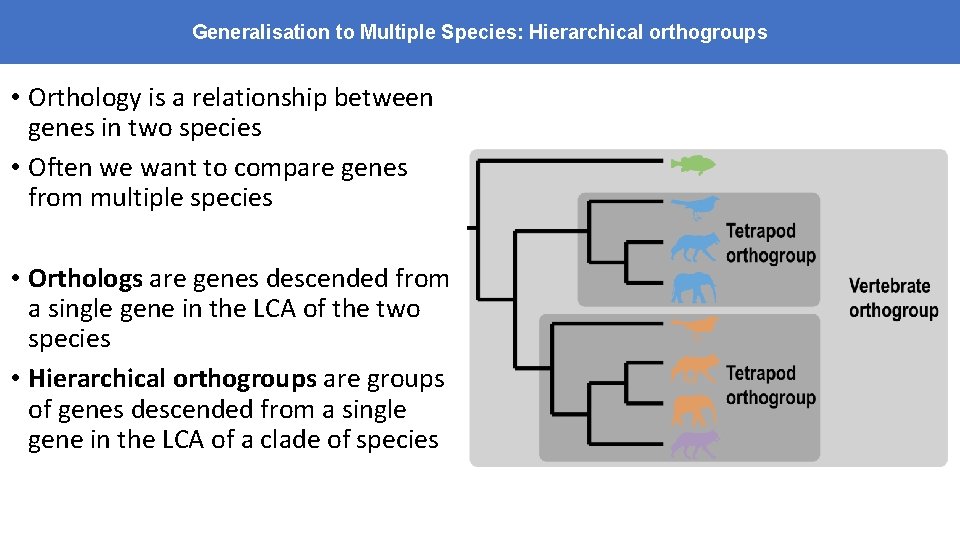
Generalisation to Multiple Species: Hierarchical orthogroups • Orthology is a relationship between genes in two species • Often we want to compare genes from multiple species • Orthologs are genes descended from a single gene in the LCA of the two species • Hierarchical orthogroups are groups of genes descended from a single gene in the LCA of a clade of species

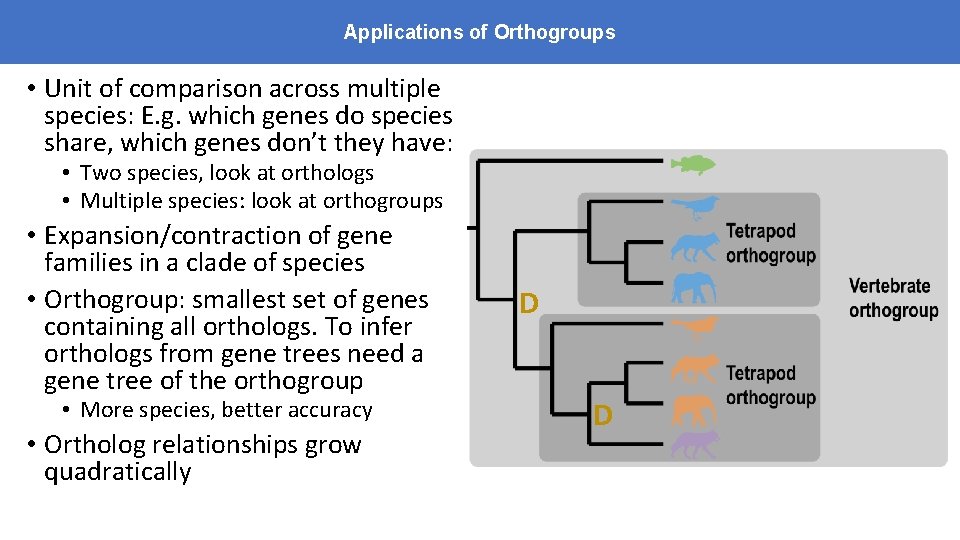
Applications of Orthogroups • Unit of comparison across multiple species: E. g. which genes do species share, which genes don’t they have: • Two species, look at orthologs • Multiple species: look at orthogroups • Expansion/contraction of gene families in a clade of species • Orthogroup: smallest set of genes containing all orthologs. To infer orthologs from gene trees need a gene tree of the orthogroup • More species, better accuracy • Ortholog relationships grow quadratically D D

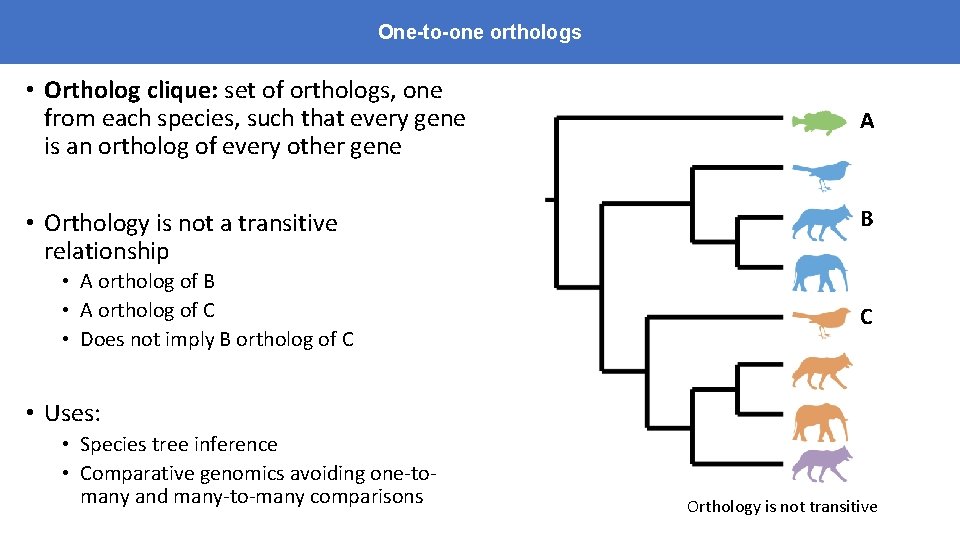
One-to-one orthologs • Ortholog clique: set of orthologs, one from each species, such that every gene is an ortholog of every other gene • Orthology is not a transitive relationship • A ortholog of B • A ortholog of C • Does not imply B ortholog of C A B C • Uses: • Species tree inference • Comparative genomics avoiding one-tomany and many-to-many comparisons Orthology is not transitive

Terminology Confusion Multiple, conflicting terms are used for the same concept. This could be a unique opportunity to bring consistency Some terminology is universally agreed: • Homolog • Ortholog • Paralog Other lesser-used but widely recognised terminology: • Co-ortholog • In-paralog, out-paralog

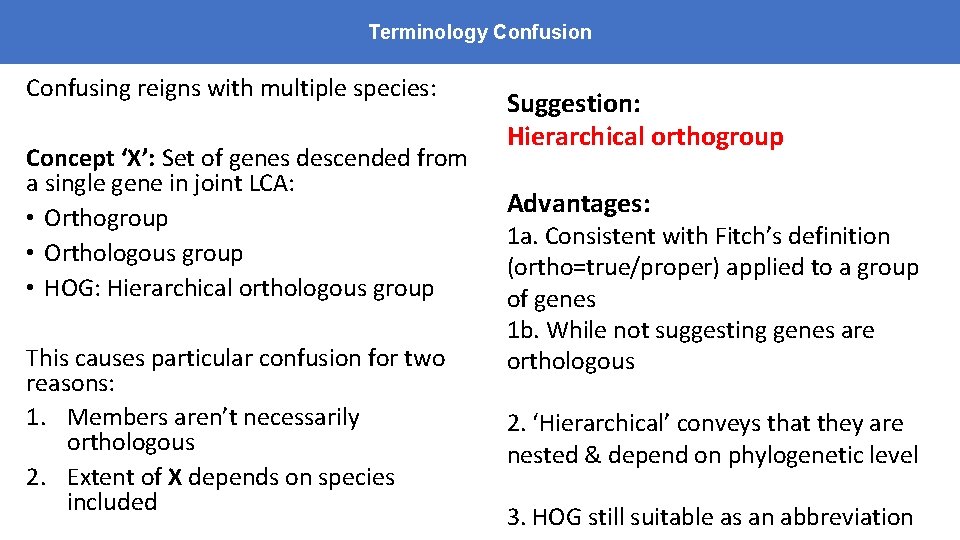
Terminology Confusion Confusing reigns with multiple species: Concept ‘X’: Set of genes descended from a single gene in joint LCA: • Orthogroup • Orthologous group • HOG: Hierarchical orthologous group This causes particular confusion for two reasons: 1. Members aren’t necessarily orthologous 2. Extent of X depends on species included Suggestion: Hierarchical orthogroup Advantages: 1 a. Consistent with Fitch’s definition (ortho=true/proper) applied to a group of genes 1 b. While not suggesting genes are orthologous 2. ‘Hierarchical’ conveys that they are nested & depend on phylogenetic level 3. HOG still suitable as an abbreviation

Terminology Confusion Confusing reigns with multiple species: Concept ‘Y’: Set of genes which are all orthologs of one another • Cliques of ortholog pairs • OMA Groups • (One-to-one orthologs) Suggestion: Ortholog clique Advantages: 1. Precise terminology, term explains its meaning exactly 1. Everything is an ortholog of everything else

Ortholog Inference Methods

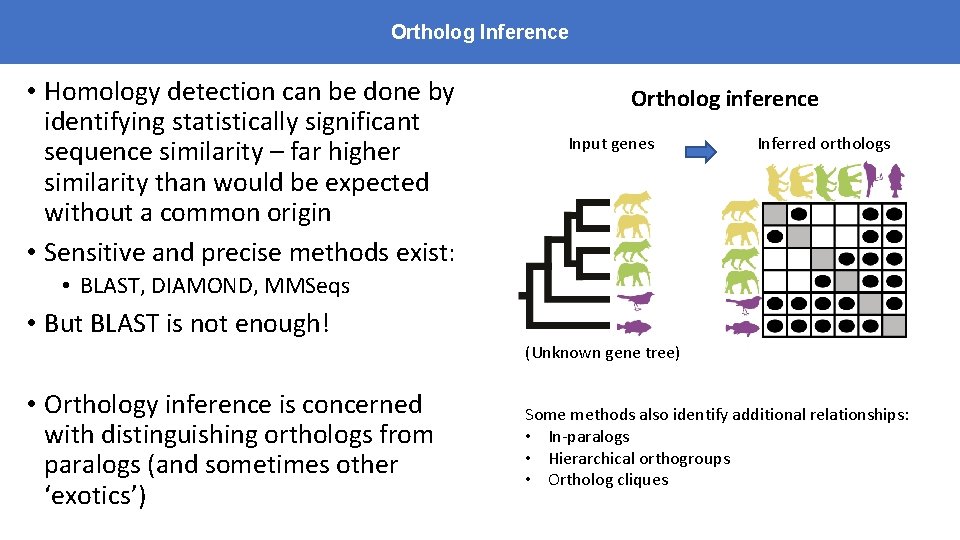
Ortholog Inference • Homology detection can be done by identifying statistically significant sequence similarity – far higher similarity than would be expected without a common origin • Sensitive and precise methods exist: Ortholog inference Input genes Inferred orthologs • BLAST, DIAMOND, MMSeqs • But BLAST is not enough! (Unknown gene tree) • Orthology inference is concerned with distinguishing orthologs from paralogs (and sometimes other ‘exotics’) Some methods also identify additional relationships: • In-paralogs • Hierarchical orthogroups • Ortholog cliques

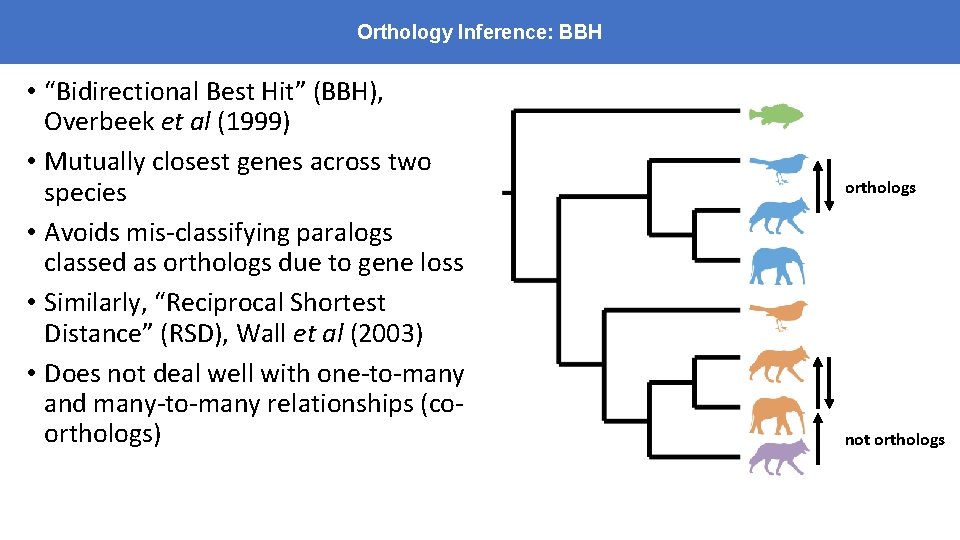
Orthology Inference: BBH • “Bidirectional Best Hit” (BBH), Overbeek et al (1999) • Mutually closest genes across two species • Avoids mis-classifying paralogs classed as orthologs due to gene loss • Similarly, “Reciprocal Shortest Distance” (RSD), Wall et al (2003) • Does not deal well with one-to-many and many-to-many relationships (coorthologs) orthologs not orthologs

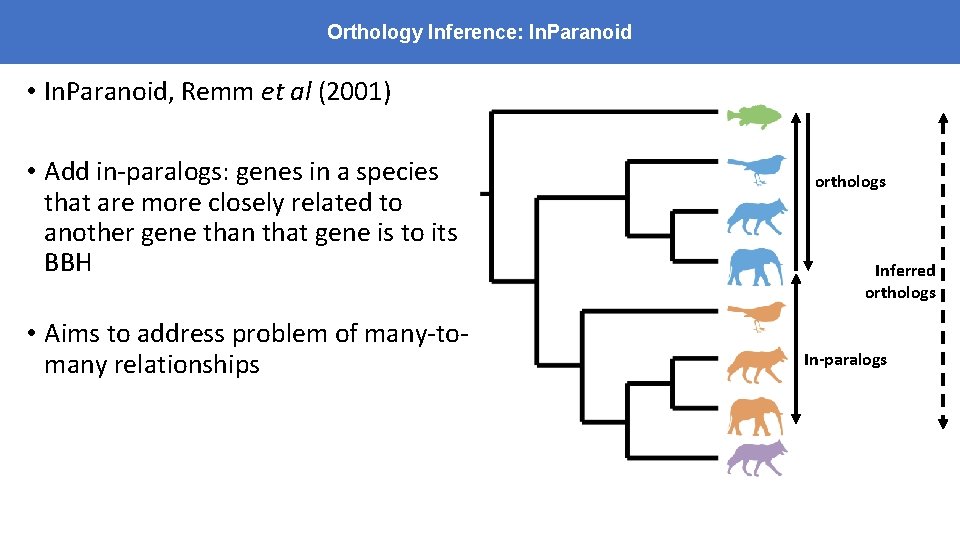
Orthology Inference: In. Paranoid • In. Paranoid, Remm et al (2001) • Add in-paralogs: genes in a species that are more closely related to another gene than that gene is to its BBH • Aims to address problem of many-tomany relationships orthologs Inferred orthologs In-paralogs

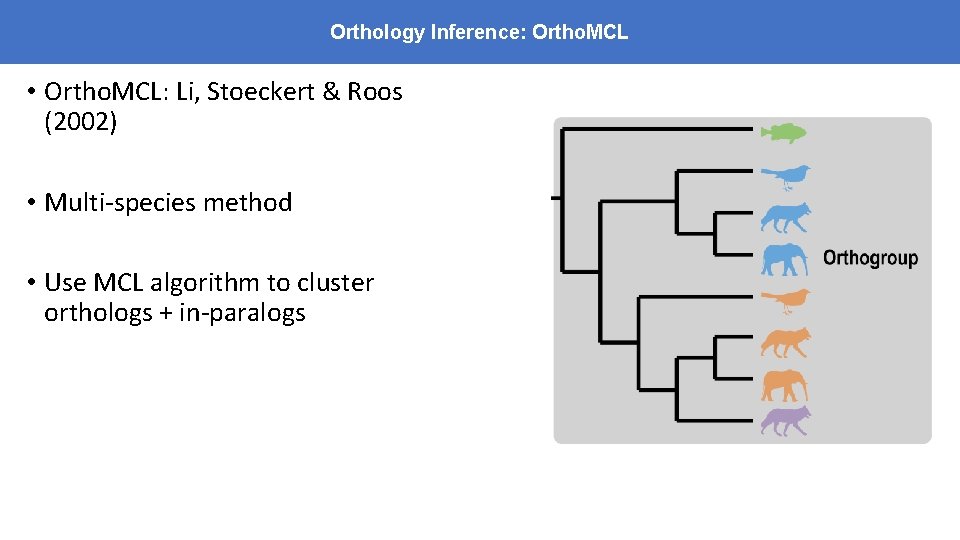
Orthology Inference: Ortho. MCL • Ortho. MCL: Li, Stoeckert & Roos (2002) • Multi-species method • Use MCL algorithm to cluster orthologs + in-paralogs

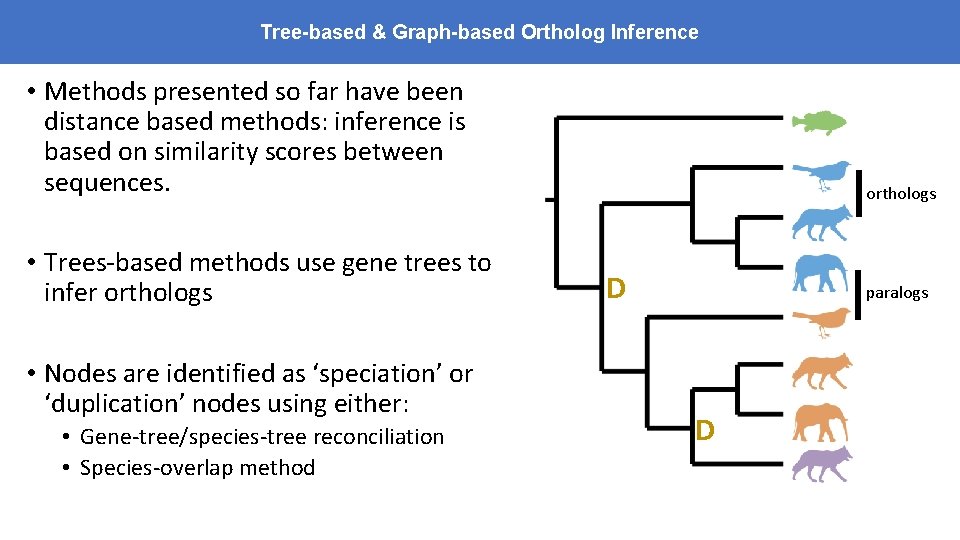
Tree-based & Graph-based Ortholog Inference • Methods presented so far have been distance based methods: inference is based on similarity scores between sequences. • Trees-based methods use gene trees to infer orthologs • Nodes are identified as ‘speciation’ or ‘duplication’ nodes using either: • Gene-tree/species-tree reconciliation • Species-overlap method orthologs D paralogs D

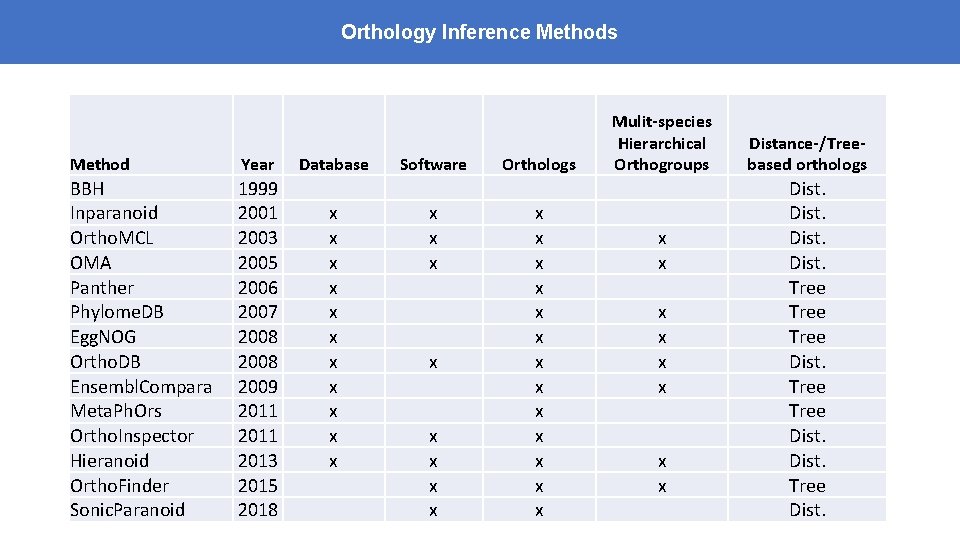
Orthology Inference Methods Method Year BBH Inparanoid Ortho. MCL OMA Panther Phylome. DB Egg. NOG Ortho. DB Ensembl. Compara Meta. Ph. Ors Ortho. Inspector Hieranoid Ortho. Finder Sonic. Paranoid 1999 2001 2003 2005 2006 2007 2008 2009 2011 2013 2015 2018 Database x x x Software x x x x Orthologs x x x x Mulit-species Hierarchical Orthogroups x x x x Distance-/Treebased orthologs Dist. Tree Dist.

Other data: Structural Conservation • Selection on proteins operates predominantly at the structural level • 3 D structure tends to exhibit higher conservation • Methods being developed for 3 D structure evolution • Tree inference from structure

Other data: Synteny • Genomes often contain blocks of conserved gene order • Conservation is more common for closely related species • Synteny has been combined with other measures in ortholog inference: SYNERGY: Wapinski, Friedman & Regev (2007)

Orthology/paralogy aren’t always applicable at gene level • Gene fusion events can result in a single gene in which different parts have different evolutionary histories • In these cases orthology and paralogy only apply at the level of the different domains, which each have their own well-defined history • Domainoid (2019)

Ortholog Benchmarks

Benchmarking Orthology Inference Methods • Given the importance of orthology there is a need for benchmarks to help: • Assess accuracy • Identify strengths and weaknesses • Guide improvements • The Quest for Orthologs consortium provides a standardized set of benchmarks with results from most methods: https: //orthology. benchmarkservice. org • Benchmarking is challenging • Hard to obtain ground-truth orthologs • Less direct approaches

Benchmarking Orthology Inference Methods • Quest for Orthologs Benchmarks: • Agreement with orthologs from human curated gene-trees (Swiss. Tree, Tree. Fam) • Species tree discordance test • a gene tree of orthologs should have the same topology as the (known) species tree • Assess methods on: • Proportion of times a samples gene has a set of orthologs for the required species/clades • Average Robison-Foulds distance between inferred gene tree and known species tree (Altenhoff et al 2016)

Benchmarking Hierarchical Orthogroup Inference Methods • Similar challenges apply to benchmarking Hierarchical Orthogroup methods • Expert curated orthogroups have been published which automated methods can be tested against (Trachana 2011) • 70 manually curated families • Range of challenges: • • Fast-/slow-evolving families Single domain vs multiple repeated domain proteins Gene duplication & loss Variable alignment quality

Applications

Applications: Gene Function Prediction • Genome Annotation: • Identify coding regions, intron-exon boundaries, functional annotation • Function prediction: • Ortholog conjecture: orthologs tend to retain their ancestral function • For 414 essential yeast genes with 1: 1 human orthologs, 43% could be replaced with their human ortholog (Kachroo et al. 2015). • Identifying candidate genes: Medical, agricultural • Different uses: • Orthologs • Homologs • Hierarchical orthogroups

Applications: Species Tree Inference • Species tree inference originally performed with morphological characters • Now almost entirely with sequence data • Marker genes 16 S r. RNA • Any orthologous genes • Deep branches hard to resolve with single genes • Data from 100 s-1000 s of loci are used • Supermatrix method: sequence data is combined and used for tree inference • Supertree method: individual gene trees are inferred and used to infer the species tree • Challenges: Incomplete lineage sorting

Applications: Reconstructing genome evolution • Hierachical orthogroups can be used to reconstruct the history of those genes: • Gene duplication & loss events • Lateral gene transfer • Changes can be pin-pointed to a branch on the species tree, complete set of changes on a branch can be inferred • Ancestral genome reconstruction: • Gene content • Protein sequences

References • Fitch, Distinguishing Homologous from Analogous Proteins, Systematic Biology, 19: 2, 1970 • Koonin, Orthologs, Paralogs, and Evolutionary Genomics, Annual Review of Genetics 39: 1, 2005 • Glover, Dessimoz, et al, Advances and Applications in the Quest for Orthologs, Molecular Biology and Evolution, 36: 10, 2019 • Darby et al, Xenolog classification, Bioinformatics, 33, : 5, 2017 • Altenhoff et al, Standardized benchmarking in the quest for orthologs, Nature Methods, 13: 425, 2016 • Trachana et al Orthology prediction methods: a quality assessment using curated protein families, Bioessays 33: 10 2011 • Kuzniar et al, The quest for orthologs: finding the corresponding gene across genomes, Trends in Genetics 24: 11 2008

References • Kachroo et al. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348: 6237, 2015

Thank you Christophe Dessimoz & Lab Steve Kelly & Lab Quest for Orthologs Consortium

New Model Species are being Used to Investigate a Wide Range of Traits • With advances in genome sequencing, easier to work with wider range of species • Investigate new traits E. g. • Aging • C 4 photosynthesis Need to: • Transfer knowledge gained back to other organisms • But also transfer to these new model organisms what we already know