Why shall we enrich proteins with specific isotopes








- Slides: 46

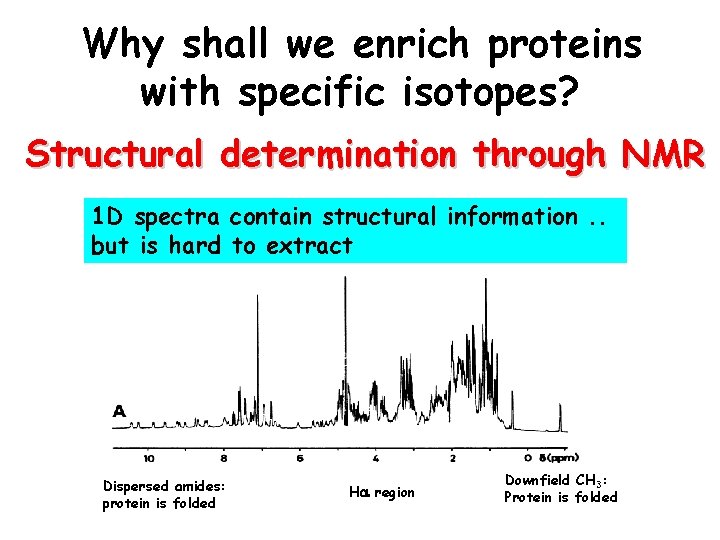
Why shall we enrich proteins with specific isotopes? Structural determination through NMR 1 D spectra contain structural information. . but is hard to extract Dispersed amides: protein is folded Ha region Downfield CH 3: Protein is folded

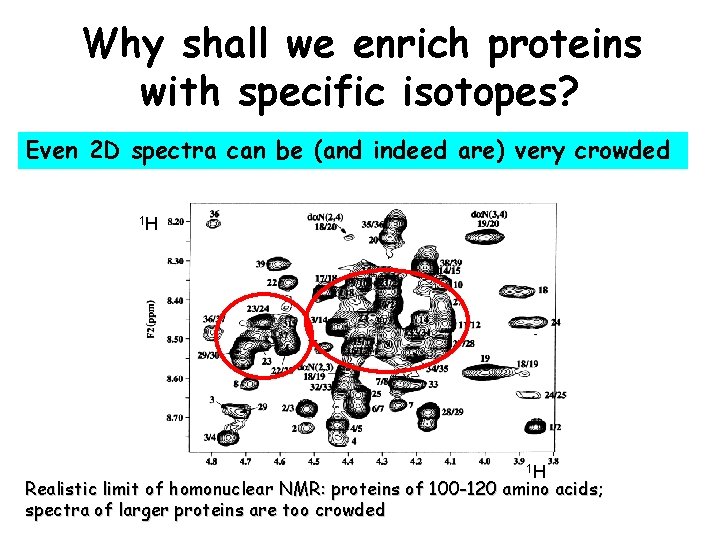
Why shall we enrich proteins with specific isotopes? Even 2 D spectra can be (and indeed are) very crowded 1 H 1 H Realistic limit of homonuclear NMR: proteins of 100 -120 amino acids; spectra of larger proteins are too crowded

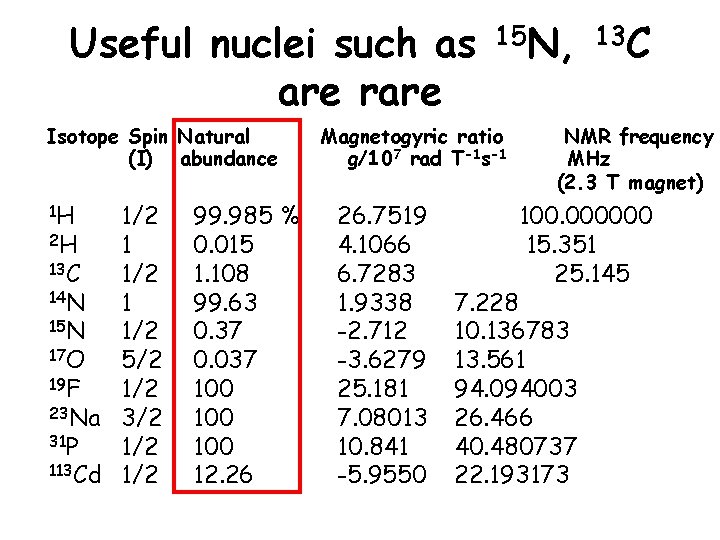
Useful nuclei such as are rare Isotope Spin Natural (I) abundance 1 H 1/2 2 H 1 13 C 1/2 14 N 1 15 N 1/2 17 O 5/2 19 F 1/2 23 Na 3/2 31 P 1/2 113 Cd 1/2 99. 985 % 0. 015 1. 108 99. 63 0. 37 0. 037 100 100 12. 26 15 N, 13 C Magnetogyric ratio g/107 rad T-1 s-1 26. 7519 4. 1066 6. 7283 1. 9338 -2. 712 -3. 6279 25. 181 7. 08013 10. 841 -5. 9550 NMR frequency MHz (2. 3 T magnet) 100. 000000 15. 351 25. 145 7. 228 10. 136783 13. 561 94. 094003 26. 466 40. 480737 22. 193173

The solution is… The solution is 3 D heteronuclear NMR Isotopic labeling

Requirements for heteronuclear NMR: isotope labeling Uniform labeling Isotopically labeled proteins can be prepared straightforwardly in E. coli by growing cells in minimal media (e. g. M 9) supplemented with appropriate nutrients (15 NH 4 Cl, 13 C-glucose) or in labelled media. Residue specific labeling Metabolic pathways can be exploited and/or appropriate auxotrophic strains of E. coli can also be used for residue selective labeling

Requirements for heteronuclear NMR: isotope labeling Deuterium labeling For large proteins deuterium labeling provides simplified spectra for the remaining 1 H nuclei and has useful effects on relaxation properties of attached or adjacent atoms (1 H, 15 N, 13 C). -Fractional and complete deuteration Labeling in eukaryotic organisms Eukaryotic proteins which are inefficiently expressed in bacteria can be efficiently expressed and labelled in yeast strains (P. pastoris)

Isotopic Uniform Labelling of proteins 15 N-labelled Protein Preparation The protein is produced by expression from bacteria which are grown on minimal medium supplemented with 15 NH 4 Cl and wild-type (wt) glucose. 15 N, 13 C-labelled Protein Preparation 15 N, 13 C-labelling is commonly referred to as double labelling. The protein is produced by expression from bacteria which are grown on minimal medium supplemented with 15 NH 4 Cl and 13 C-glucose. 15 N, 13 C, 2 H-labelled Protein Preparation 15 N, 13 C, 2 H-labelling is commonly referred to as triple labelling. The protein is produced by expression from bacteria which are grown on minimal medium supplemented with 15 NH 4 Cl and 13 C-glucose and using D 2 O instead of H 2 O. This will result in about 70 -80% deuteration of the side-chains, as there is a certain amount of contaminating 1 H present from the glucose. Higher levels of deuteration of around 95% can be achieved if 13 C, 2 H-glucose is used. However, this is more expensive and in many cases the cheaper version is sufficient. Note that the NH groups are exchangeable. This means that they will back-exchange to 1 H when the protein is purified in normal aqueous solution. In this way, many of the normal NHbased experiments can be carried out on triple-labelled protein.

Sources of isotopes used for uniform labeling In most cases (except in cell-free labelling), the protein is expressed by bacteria. The isotopic labels are introduced by feeding the bacteria specific nutrients. In most cases the basis will be so-called minimal medium which contains all the salts and trace elements needed by the bacteria but contains no carbon or nitrogen sources. These elements can then be introduced using a variety of different isotopically labelled carbon and nitrogen sources. The simplest labeling, and also the cheapest one is 15 N, because 15 N ammonia is quite cheap. 13 C is more expensive, because it requires the synthesis (most commonly - biosynthesis) of 13 C glucose or glycerol. Some expression systems allow use of cheaper 13 CO 2.

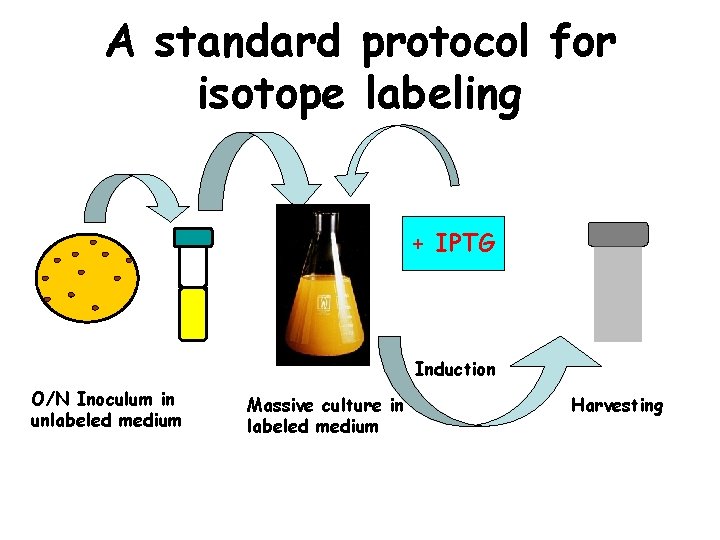
A standard protocol for isotope labeling + IPTG Induction O/N Inoculum in unlabeled medium Massive culture in labeled medium Harvesting

From protein purification to check folding Protein isolation and purification will follow the standard procedure which has been set up for unlabelled protein Check folding Protein folding can be checked by 1 H NMR and 1 H-15 N HSQC spectra.

How to optimize protein expression? Choice of culture medium Two main types of culture media can be tested for uniform labeling: Ready-to-use media like algae or bacteria hydrolysate Minimal media added with 15 N nitrogen source or/and 13 C carbon source

Minimal media are composed in the lab and are made of nutrients like C and N source, salts, buffering substances, traces elements and vitamins. Carbon source can be glucose (the best as gives highest yields), glycerol, acetate, succinate, methanol, Etc. In case of 13 C labeling the concentration of carbon source can be reduced with respect to unlabelled culture, to reduce costs!!! Checks must be performed before labelling! Nitrogen source can be NH 4 Cl or (NH 4) 2 SO 4 In case of 15 N labeling the concentration of nitrogen source can be reduced with respect to unlabeled culture, to reduce costs!!! Checks must be performed before labeling!

Minimal media are composed in the lab and are made of nutrients like C and N source, salts, buffering substances, traces elements and vitamins. Salts are Na. Cl/KCl, Mg. SO 4, Ca. Cl 2 Buffer usually is phosphate, p. H 7. 5 Trace elements is constituted by a mixtures of metal ions, like Co 2+, Cu 2+, Zn 2+, Mn 2+, Fe 2+ Vitamins are thiamine, biotin, folic acid, niacinamide, pantothenic acid, pyridoxal, riboflavin

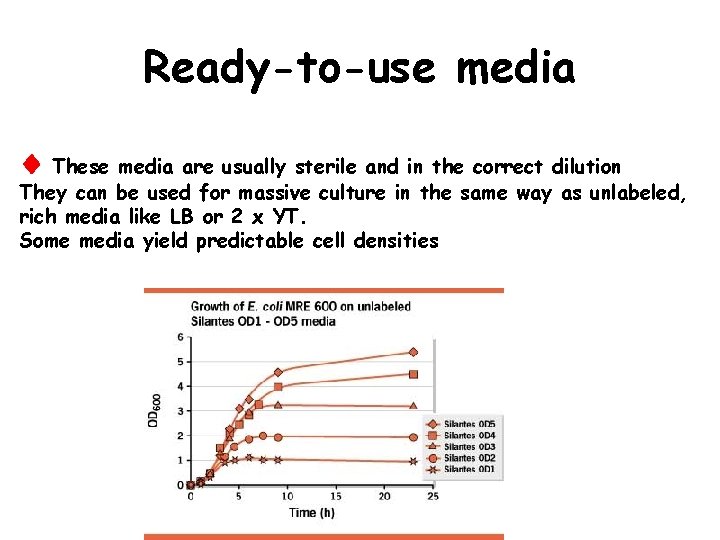
Ready-to-use media These media are usually sterile and in the correct dilution They can be used for massive culture in the same way as unlabeled, rich media like LB or 2 x YT. Some media yield predictable cell densities

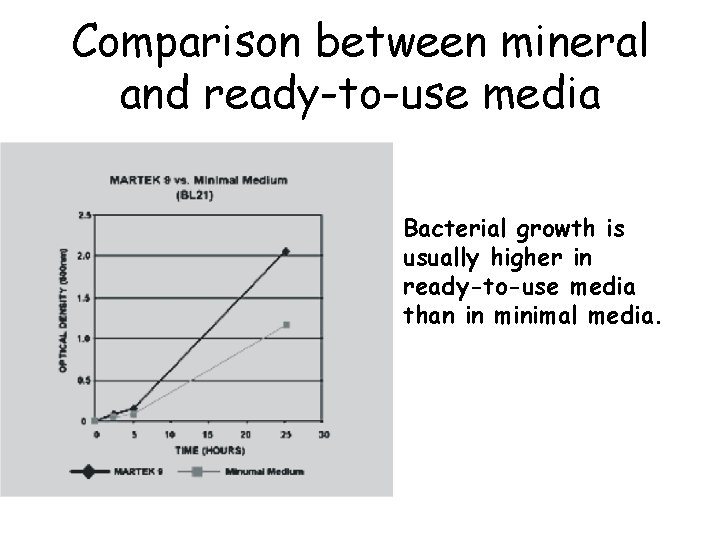
Comparison between mineral and ready-to-use media Bacterial growth is usually higher in ready-to-use media than in minimal media.

Comparison between mineral and ready-to-use media But protein expression? It must be tested, case by case, through expression tests Example:

Strategies to improve protein expression An example: Grow cell mass on unlabeled rich media allowing rapid growth to high cell density. Exchange the cell into a labeled medium at higher cell densities optimized for maximal protein expression Marley J et al. J. Biomol. NMR 2001, 20, 71 -75

Strategies to improve protein expression In practice: Cells are grown in rich unlabeled medium. When OD 600 = 0. 7 cells are harvested, washed with M 9 salt solution, w. o. N and C source and resuspended in labeled media at a higher cell concentration. Protein expression is induced after 1 hour by addition of IPTG.

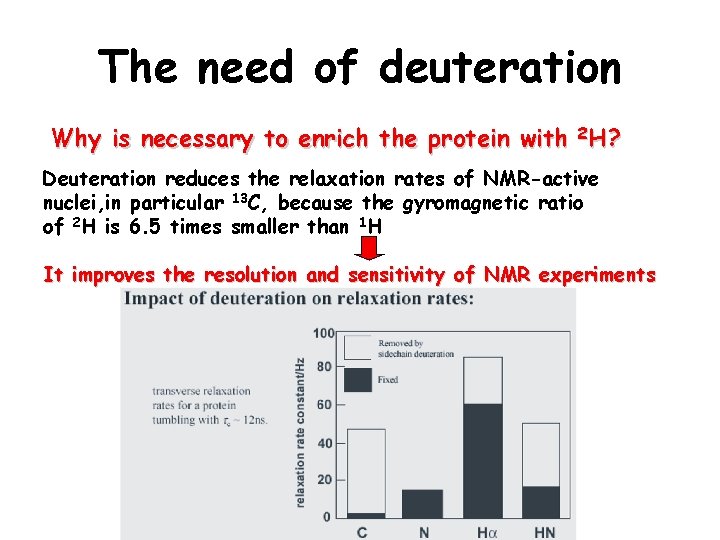
The need of deuteration Why is necessary to enrich the protein with 2 H? Deuteration reduces the relaxation rates of NMR-active nuclei, in particular 13 C, because the gyromagnetic ratio of 2 H is 6. 5 times smaller than 1 H It improves the resolution and sensitivity of NMR experiments

Which is the ideal level of deuteration? It depends from the size of the protein In general for c up to 12 ns (20 KDa) 13 C/15 N labeling for c up to 18 ns (35 KDa) 13 C/15 N labeling and fractional deuteration for c above 18 ns 13 C/15 N labeling – selective protonation and background deuteration It depends from the type of NMR experiments

The problem to express a deuterated protein Incorporation of 2 H reduces growth rate of organisms (up to 50%) and decreases protein production as a consequence of the isotopic effect. Changing a hydrogen atom to deuterium represents a 100% increase in mass, whereas in replacing carbon-12 with carbon-13, the mass increases by only 8%. The rate of a reaction involving a C–H bond is typically 6– 10 Deuterium labeling requires conditions different 12 times faster than the corresponding C–D bond, whereas a C reaction is 13 C reaction with respect to 13 C and 15 N enrichment and could only ~1. 04 times faster than the corresponding require bacteria adaptation

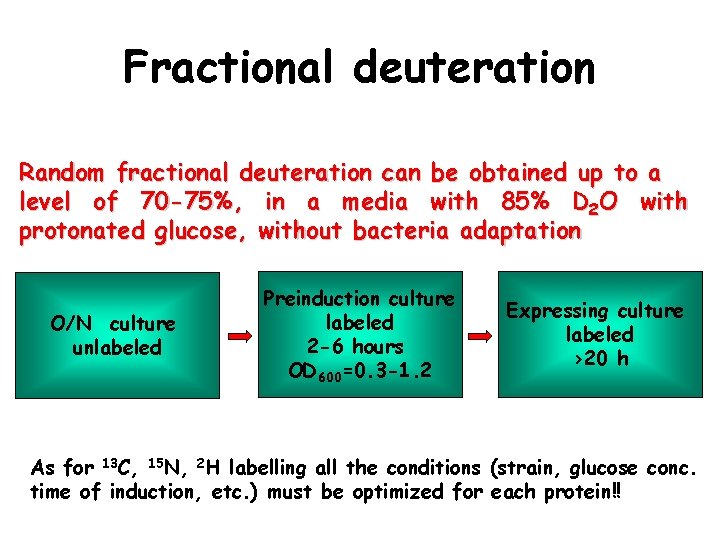
Fractional deuteration Random fractional deuteration can be obtained up to a level of 70 -75%, in a media with 85% D 2 O with protonated glucose, without bacteria adaptation O/N culture unlabeled Preinduction culture labeled 2 -6 hours OD 600=0. 3 -1. 2 Expressing culture labeled >20 h As for 13 C, 15 N, 2 H labelling all the conditions (strain, glucose conc. time of induction, etc. ) must be optimized for each protein!!

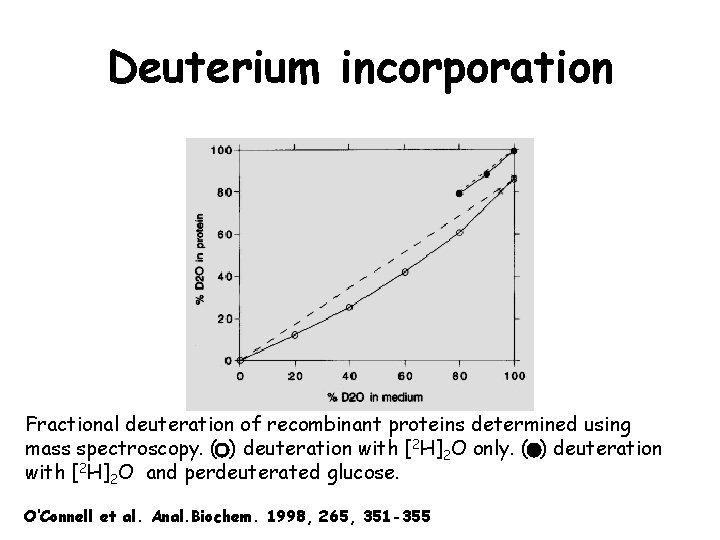
Deuterium incorporation Fractional deuteration of recombinant proteins determined using mass spectroscopy. ( ) deuteration with [2 H]2 O only. ( ) deuteration with [2 H]2 O and perdeuterated glucose. O’Connell et al. Anal. Biochem. 1998, 265, 351 -355

Perdeuteration can require a gradual adaptation of bacteria to increasing concentration of D 2 O. Bacterial strains must be accurately selected in order to choose that which better acclimates to D 2 O media. For each strain one or more colony must be selected which better survives in high level of D 2 O concetration

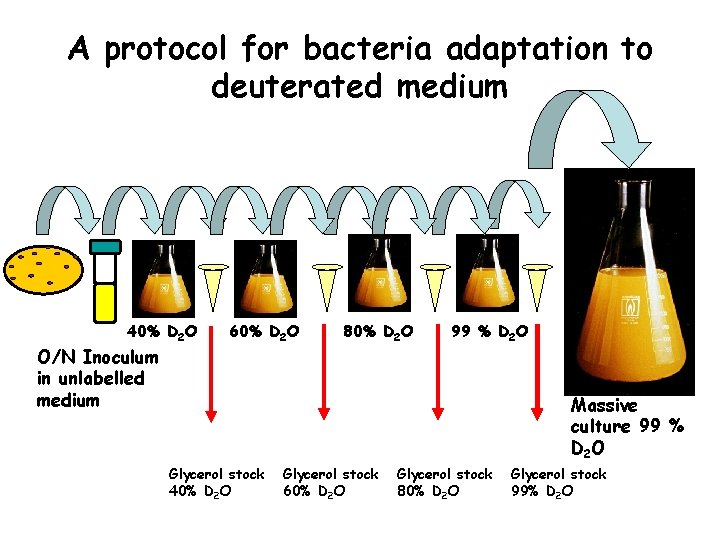
A protocol for bacteria adaptation to deuterated medium 40% D 2 O O/N Inoculum in unlabelled medium 60% D 2 O 80% D 2 O 99 % D 2 O Massive culture 99 % D 2 O Glycerol stock 40% D 2 O Glycerol stock 60% D 2 O Glycerol stock 80% D 2 O Glycerol stock 99% D 2 O

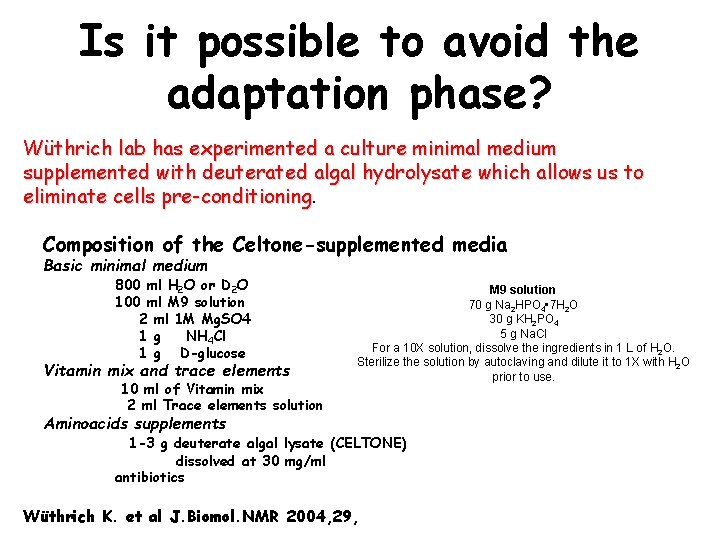
Is it possible to avoid the adaptation phase? Wüthrich lab has experimented a culture minimal medium supplemented with deuterated algal hydrolysate which allows us to eliminate cells pre-conditioning Composition of the Celtone-supplemented media Basic minimal medium 800 ml H 2 O or D 2 O 100 ml M 9 solution 2 ml 1 M Mg. SO 4 1 g NH 4 Cl 1 g D-glucose Vitamin mix and trace elements 10 ml of Vitamin mix 2 ml Trace elements solution M 9 solution 70 g Na 2 HPO 4 • 7 H 2 O 30 g KH 2 PO 4 5 g Na. Cl For a 10 X solution, dissolve the ingredients in 1 L of H 2 O. Sterilize the solution by autoclaving and dilute it to 1 X with H 2 O prior to use. Aminoacids supplements 1 -3 g deuterate algal lysate (CELTONE) dissolved at 30 mg/ml antibiotics Wüthrich K. et al J. Biomol. NMR 2004, 29,

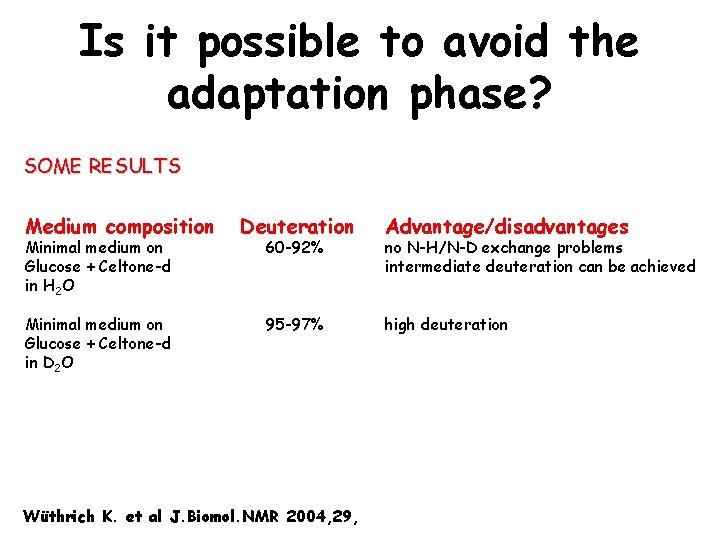
Is it possible to avoid the adaptation phase? SOME RESULTS Medium composition Minimal medium on Glucose + Celtone-d in H 2 O Minimal medium on Glucose + Celtone-d in D 2 O Deuteration 60 -92% 95 -97% Wüthrich K. et al J. Biomol. NMR 2004, 29, Advantage/disadvantages no N-H/N-D exchange problems intermediate deuteration can be achieved high deuteration

Backbone HN Side-chians

Specific labeling In Labeling of a protein can be easily achieved on specific residues with 2 strategies: a medium containing small amounts of glucose (13 C labelled or unlabelled)/NH 4 Cl (15 N labelled or unlabelled) and complemented with the labelled aminoacid(s). A mixture of the other unlabeled aminoacid(s) can be added to prevent any conversion of the labeled aminoacid(s) In a complete labelled medium, containing great amount of all unlabeled aminoacids except those which are expected to be labeled.

Specific labeling: the main problem The most important problem encountered is the metabolic conversions of the labeled aminoacids which might occur during anabolism and/or catabolism. How to prevent this? Use an auxotrophic strain. Use a prototrophic strain with high concentration of aminoacids to inhibit some metabolic pathways. An example: Labeling of a protein with 13 C 15 N Lys can be performed in unlabeled media with high level of 13 C 15 N Lys to prevent lysine biosinthesis from aspartate conversion.

Amino Acid Specific Labelling The protein is produced by expression from bacteria which are grown on minimal medium supplemented with small amounts of 15 NH 4 Cl and 13 C-labelled glucose as well as labelled and unlabelled amino acids. The idea is that only those amino acids which are added in labelled form become labelled in the protein. Unfortunately, this may not always work as desired, since the E. coli metabolism and catabolism causes a degree of interconversion between amino acids. Thus, it is not possible to create a sample with any combination of labelled amino acids. The situation can be improved somewhat by using auxotrophic bacterial strains or incorporating enzyme inhibitors. A cheaper way of labelling only certain amino acids, often called reverse labelling, involves expression from bacteria which are grown on minimal medium supplemented with 15 NH 4 Cl and 13 C-labelled glucose as well as unlabelled amino acids. This supresses the labelling of these amino acids and only those which have not been added unlabelled will be synthesised by the bacteria using the 13 C-glucose as the carbon source. Again, a certain amount of scrambling may occur. However, if complete control over the incorporation of amino acids is required, then cell-free methods must be used.

Specific labeling for assignment of 13 C and 1 H methyl from Ile, Leu, Val Full deuteration precludes the use of NOEs for structure determination. How to overcome the problem? Reintroduction of protons by using labeled amino acids Reintroduction of protons by using methyl selectivelly protonated metabolic precursors of aliphatic amino acids or the biosyntetic precursor of the aromatic rings.

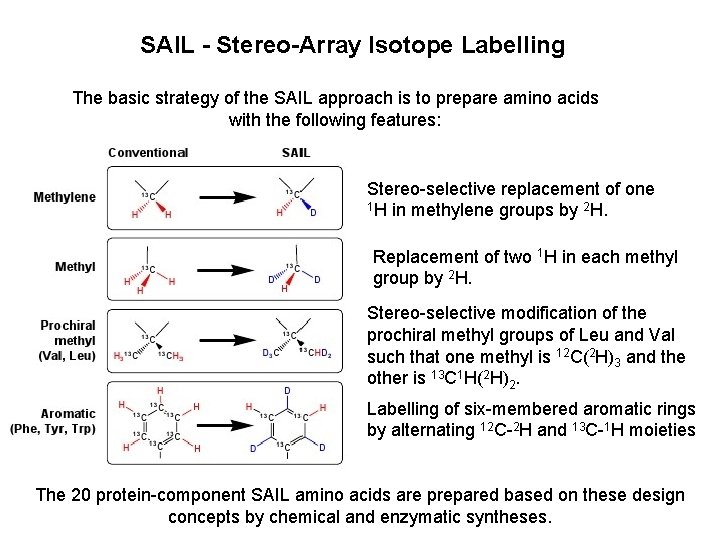
SAIL - Stereo-Array Isotope Labelling The basic strategy of the SAIL approach is to prepare amino acids with the following features: Stereo-selective replacement of one 1 H in methylene groups by 2 H. Replacement of two 1 H in each methyl group by 2 H. Stereo-selective modification of the prochiral methyl groups of Leu and Val such that one methyl is 12 C(2 H)3 and the other is 13 C 1 H(2 H)2. Labelling of six-membered aromatic rings by alternating 12 C-2 H and 13 C-1 H moieties The 20 protein-component SAIL amino acids are prepared based on these design concepts by chemical and enzymatic syntheses.

SAIL - Stereo-Array Isotope Labelling The production of SAIL proteins involves cell-free expression system. This approach indeed minimize metabolic scrambing effects and produces high incorporation rate of the added SAIL amino acid into the target protein.

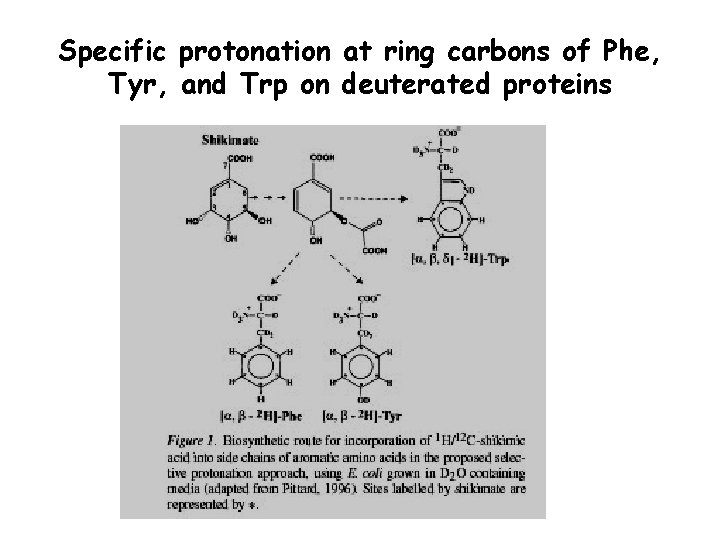
Specific protonation at ring carbons of Phe, Tyr, and Trp on deuterated proteins NOEs involving aromatic protons are an important source of distance restraints in the structure calculation of perdeuterated proteins. A selective reverse labeling of Phe, Tyr and Trp has been performed in perdeuterated proteins, using shikimic acid, a precursor of the aromatic rings. In this way the aromatic rings of the aminoacids are partially protonated (50%) Rajesh S. et al. J. Biomol. NMR 2003, 27, 81 -86

Specific protonation at ring carbons of Phe, Tyr, and Trp on deuterated proteins

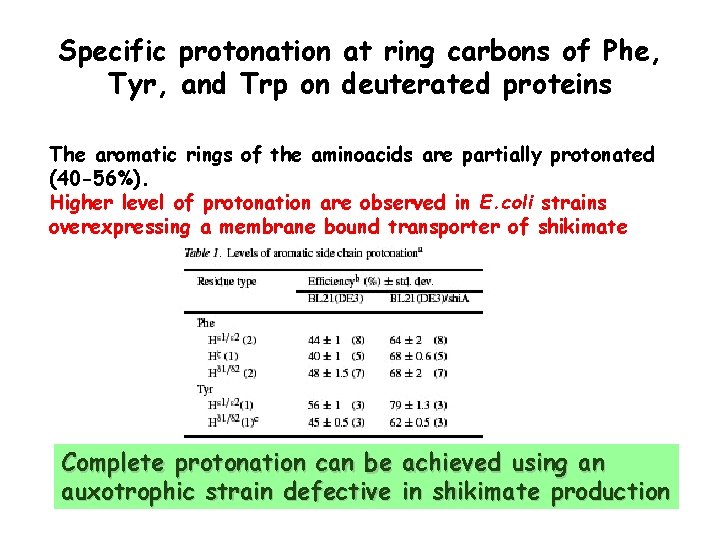
Specific protonation at ring carbons of Phe, Tyr, and Trp on deuterated proteins The aromatic rings of the aminoacids are partially protonated (40 -56%). Higher level of protonation are observed in E. coli strains overexpressing a membrane bound transporter of shikimate Complete protonation can be achieved using an auxotrophic strain defective in shikimate production

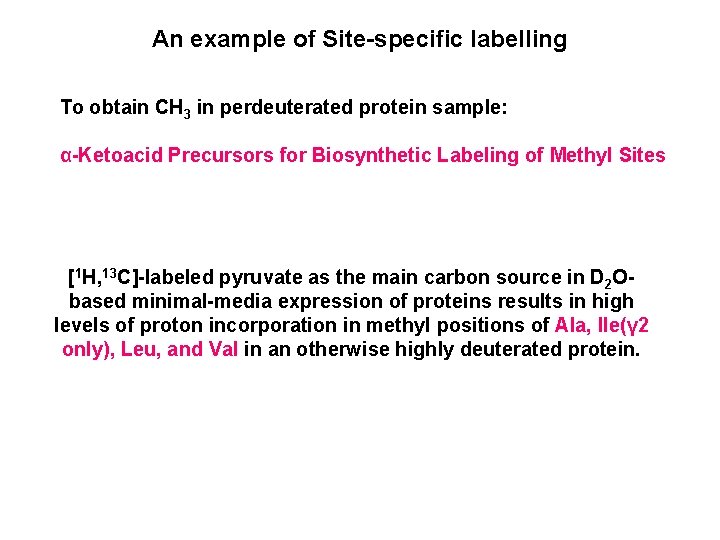
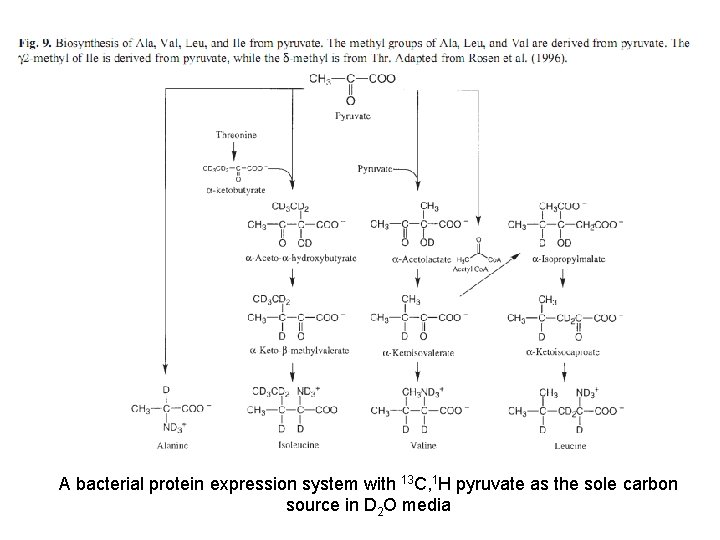
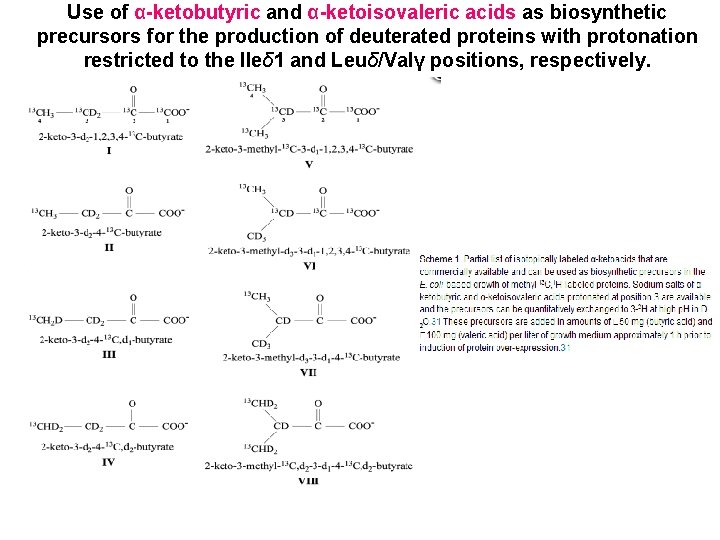
An example of Site-specific labelling To obtain CH 3 in perdeuterated protein sample: α-Ketoacid Precursors for Biosynthetic Labeling of Methyl Sites [1 H, 13 C]-labeled pyruvate as the main carbon source in D 2 Obased minimal-media expression of proteins results in high levels of proton incorporation in methyl positions of Ala, Ile(γ 2 only), Leu, and Val in an otherwise highly deuterated protein.

A bacterial protein expression system with 13 C, 1 H pyruvate as the sole carbon source in D 2 O media

Unfortunately, because the protons of the methyl group of pyruvate exchange with solvent, proteins are produced with all four of the possible methyl isotopomers (13 CH 3, 13 CH 2 D, 13 CHD 2, and 13 CD 3).

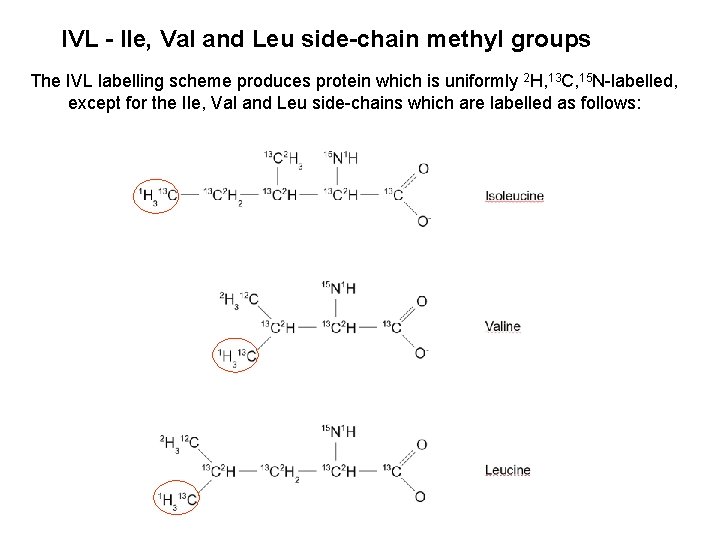
IVL - Ile, Val and Leu side-chain methyl groups The IVL labelling scheme produces protein which is uniformly 2 H, 13 C, 15 N-labelled, except for the Ile, Val and Leu side-chains which are labelled as follows:

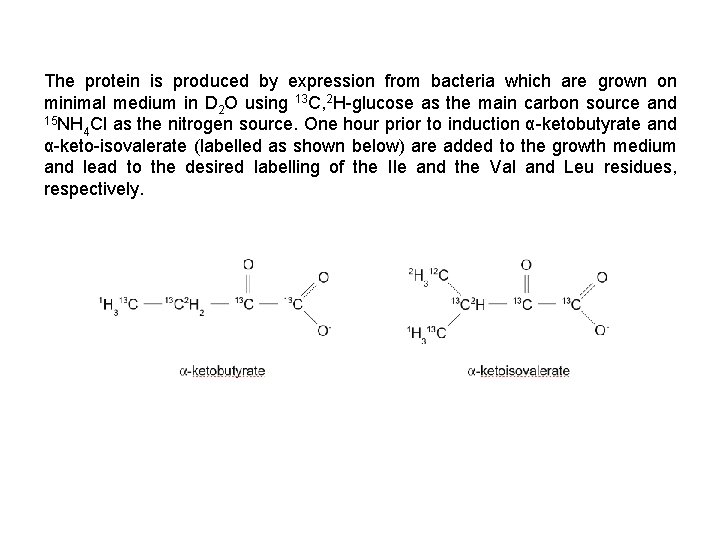
The protein is produced by expression from bacteria which are grown on minimal medium in D 2 O using 13 C, 2 H-glucose as the main carbon source and 15 NH Cl as the nitrogen source. One hour prior to induction α-ketobutyrate and 4 α-keto-isovalerate (labelled as shown below) are added to the growth medium and lead to the desired labelling of the Ile and the Val and Leu residues, respectively.

Use of α-ketobutyric and α-ketoisovaleric acids as biosynthetic precursors for the production of deuterated proteins with protonation restricted to the Ileδ 1 and Leuδ/Valγ positions, respectively.

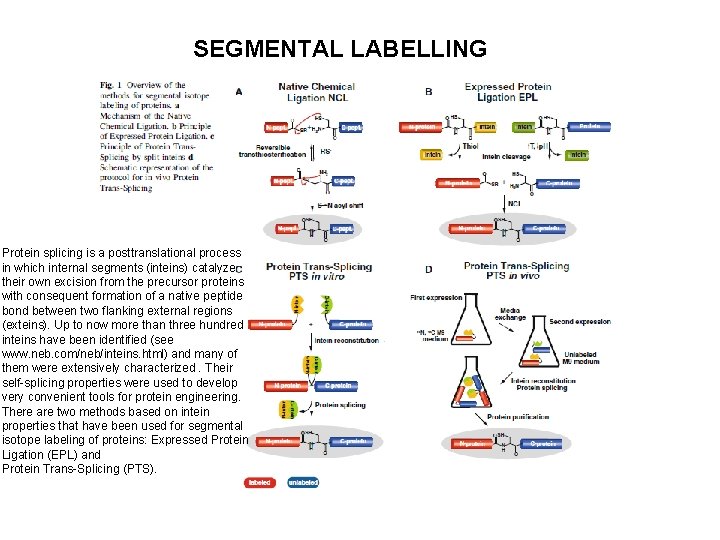
SEGMENTAL LABELLING Protein splicing is a posttranslational process in which internal segments (inteins) catalyze their own excision from the precursor proteins with consequent formation of a native peptide bond between two flanking external regions (exteins). Up to now more than three hundred inteins have been identified (see www. neb. com/neb/inteins. html) and many of them were extensively characterized. Their self-splicing properties were used to develop very convenient tools for protein engineering. There are two methods based on intein properties that have been used for segmental isotope labeling of proteins: Expressed Protein Ligation (EPL) and Protein Trans-Splicing (PTS).

METODI DI ARRICCHIMENTO ISOTOPICO Ø Uniform labeling All atoms of a selected element are represented by a single isotope Ø Partial labeling A selected element is present in a mixture of isotopic forms. It's not possible to use 15 N of the amino acid to label because cell in which we express the protein have transaminase that make fast exchange of the label. Deuterium labeling could be done only for a portion of all hydrogens. Ø Site-specific labelling In the site-specific labeling approach only certain residues, or particular atoms in some residues are isotopically labeled

Minimal media Trace Elements In 800 ml H 2 O dissolve 5 g Na 2 EDTA and correct to p. H 7 Add the following in order, correcting to p. H 7 after each: Fe. Cl 3 (. 6 H 20) 0. 5 g (0. 83 g) Zn. Cl 2 0. 05 g Cu. Cl 2 0. 01 g Co. Cl 2. 6 H 2 O 0. 01 g H 3 BO 3 0. 01 g Mn. Cl 2. 6 H 2 O (. 4 H 20) 1. 6 g (1. 35 g) Make up to 1 litre, autoclave and store at 4°C. M 9 -minimal media: Per litre, adds: 7 g Na 2 HPO 4 3 g KH 2 PO 4 0. 5 g Na. Cl M 9 -Solution Then add: 1 ml 1 M Mg. SO 4 200 µl 1 M Ca. Cl 2 1 ml Thiamine (40 mg ml-1 stock) 10 ml Trace Elements Also add, as necessary: 15 ml Glucose (20 % Stock) (gives 0. 3 % final) 1 g NH 4 Cl