WholeGenome Prokaryote Phylogeny without Sequence Alignment Bailin HAO
Whole-Genome Prokaryote Phylogeny without Sequence Alignment Bailin HAO and Ji QI T-Life Research Center, Fudan University Shanghai 200433, China Institute of Theoretical Physics, Academia Sinica Beijing 100080, China http: //www. itp. ac. cn/~hao/
Classification of Prokaryotes: A Long-Standing Problem • Traditional taxonomy: too few features • Morphology:spheric, helices, rod-shaped…… • Metabolism:photosythesis, N-fixing, desulfurization…… • Gram staining:positive and negative • SSU r. RNA Tree (Carl Woese et al. , 1977): – 16 S r. RNA: ancient conserved sequences of about 1500 kb – Discovery of the three domains of life: Archaea, Bacteria and Eucarya – Support to endosymbiont origin of mitochondria and chloroplasts

The SSU r. RNA Tree of Life: A big progress in molecular phylogeny of prokaryotes as evidenced by the history of the Bergey’s Manual

Bergey’s Manual Trust: Bergey’s Manual • 1 st Ed. “Determinative Bacteriology”: 1923 • 8 th Ed. “Determinative Bacteriology”: 1974 • 1 st Ed. “Systematic Bacteriology”: 1984 -1989, 4 volumes • 9 th Ed. “Determinative Bacteriology”: 1994 • 2 nd Ed. “Systematic Bacteriology”: 2001 -200? , 5 volumes planned; On-Line “Taxonomic Outline of Procarytes” by Garrity et al. Rel. 4. 0 (October 2003): 26 phyla: A 1 -A 2, B 1 -B 24
Phylogeny versus Taxonomy • Phylogeny and taxonomy are not synonyms • Taxonomy – classification, systematics of extant species • Phylogeny – the history of evolution since the origin of species • One should not contradict the two with each other • From the Preface to Outline of Procaryotes (Rel. 4. 0, October 2003): “The primary objective was to devise a classification that would reflect the phylogeny of procaryotes, …”

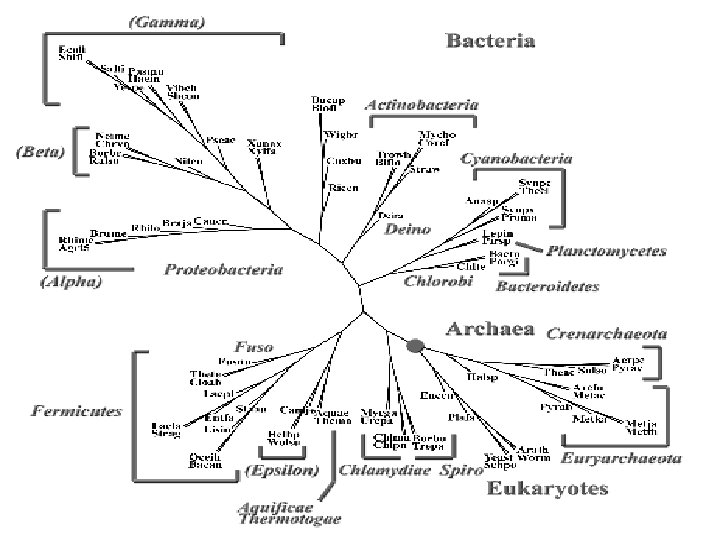
Our Latest Result • • • NCBI Genome data as of 31 December 2004 222 organisms = (21 A + 193 B + 8 E) Input: genome data (the. faa files) Output: a phylogenetic tree No selection of genes, no alignment of sequences, no fine adjustment whatsoever • See the tree first. Story follows.

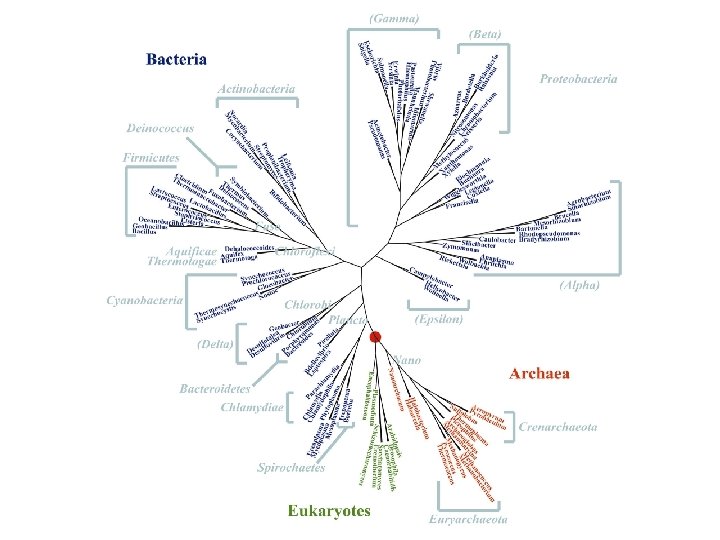
Complete Bacterial Genomes Appeared since 1995 Early Expectations: • More support to the SSU r. RNA Tree of Life • Add details to the classification (branchings and groupings) • More hints on taxonomic revisions
Confusion brought by the hyperthermophiles – Aquifex aeolicus (Aquae) 1998: 1551335 – Thermotoga maritima (Thema) 1999: 1860725 – “Genome Data Shake tree of life” Science 280 (1 May 1998) 672 – “Is it time to uproot the tree of life? ” Science 284 (21 May 1999) 130 – “Uprooting the tree of life” W. Ford Doolittle, Scientific American (February 2000) 90
Debate on Lateral Gene Transfer • Extreme estimate: 17% in E. Coli Limitations of the above approach B. Wang, J. Mol. Evol. 53 (2001) 244 • “Phase transition” and “crystalization” of species (C. Woese 1998) • Lateral transfer within smaller gene pools as an innovative agent • Composition vector may incorporate LGT within small gene pools
Our Motivations: • Develop a molecular phylogeny method that makes use of complete genomes – no selection of particular genes • Avoid sequence alignment • Try to reach higher resolution to provide an independent comparison with other approaches such as SSU r. RNA trees • Make comparison with bacteriologists’ systematics as reflected in Bergey’s Manual (2001 - 2003) • Qi, Wang, Hao, J. Molecular Evolution, 58 (1) (Jaunary 2004) 1 – 11. (109=16 A+87 B+6 E)
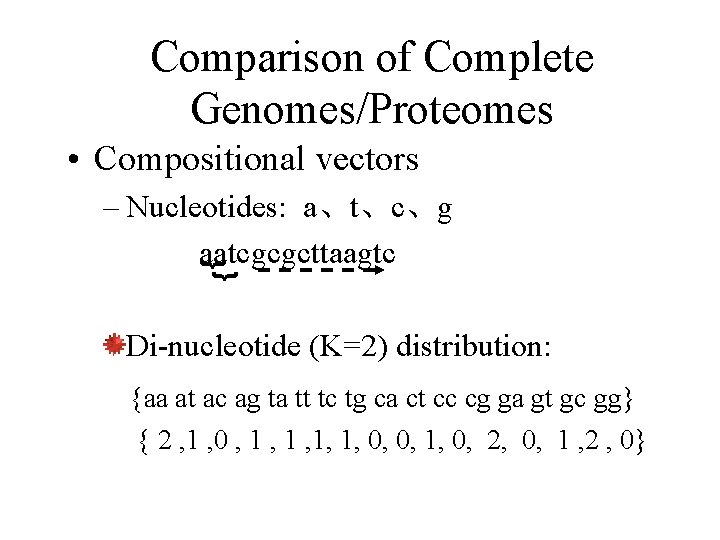
Comparison of Complete Genomes/Proteomes • Compositional vectors }} – Nucleotides: a、t、c、g aatcgcgcttaagtc Di-nucleotide (K=2) distribution: {aa at ac ag ta tt tc tg ca ct cc cg ga gt gc gg} { 2 , 1 , 0 , 1 , 1, 1, 0, 0, 1, 0, 2, 0, 1 , 2 , 0}
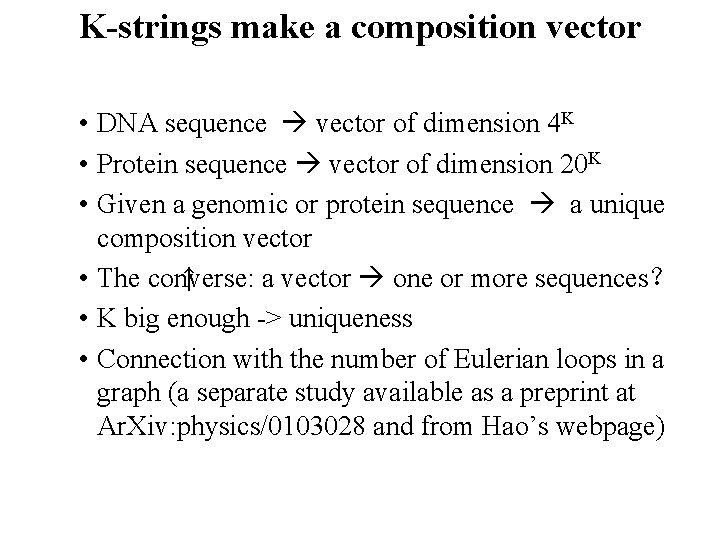
K-strings make a composition vector • DNA sequence vector of dimension 4 K • Protein sequence vector of dimension 20 K • Given a genomic or protein sequence a unique composition vector ↑ • The converse: a vector one or more sequences? • K big enough -> uniqueness • Connection with the number of Eulerian loops in a graph (a separate study available as a preprint at Ar. Xiv: physics/0103028 and from Hao’s webpage)
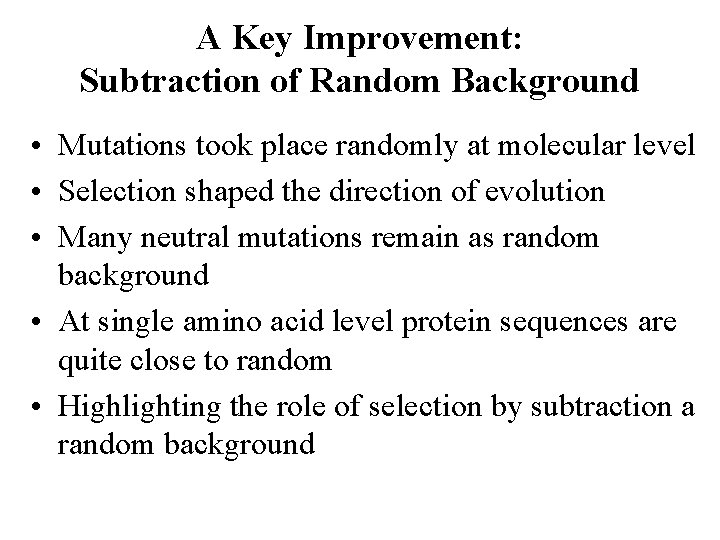
A Key Improvement: Subtraction of Random Background • Mutations took place randomly at molecular level • Selection shaped the direction of evolution • Many neutral mutations remain as random background • At single amino acid level protein sequences are quite close to random • Highlighting the role of selection by subtraction a random background
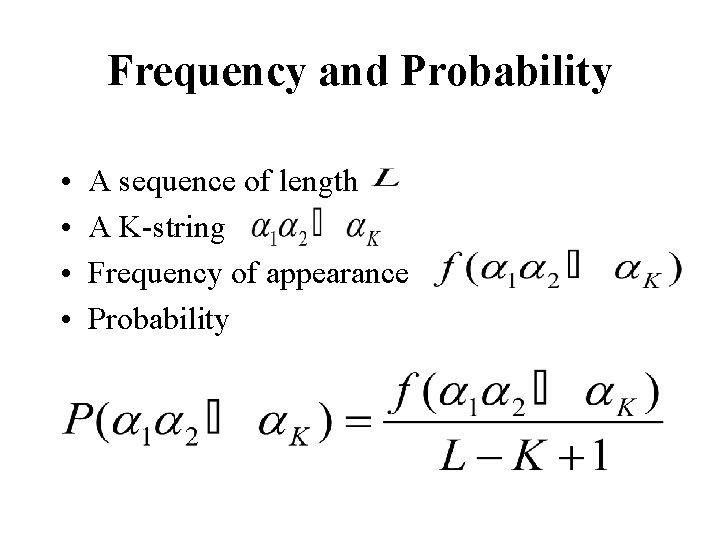
Frequency and Probability • • A sequence of length A K-string Frequency of appearance Probability
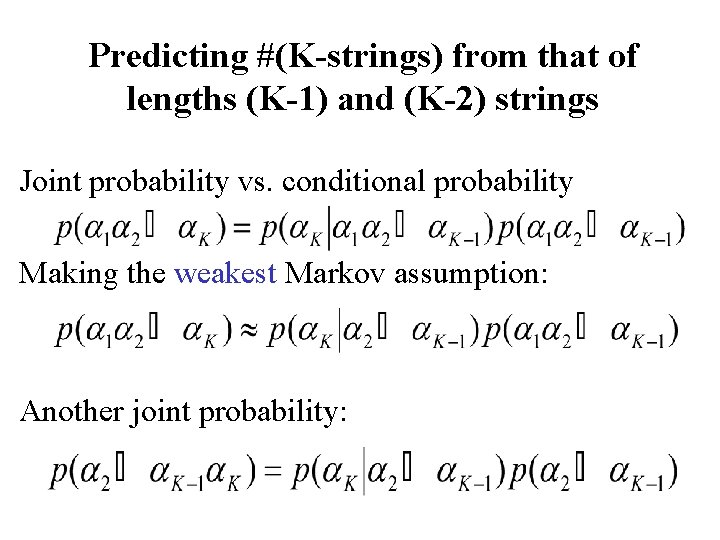
Predicting #(K-strings) from that of lengths (K-1) and (K-2) strings Joint probability vs. conditional probability Making the weakest Markov assumption: Another joint probability:
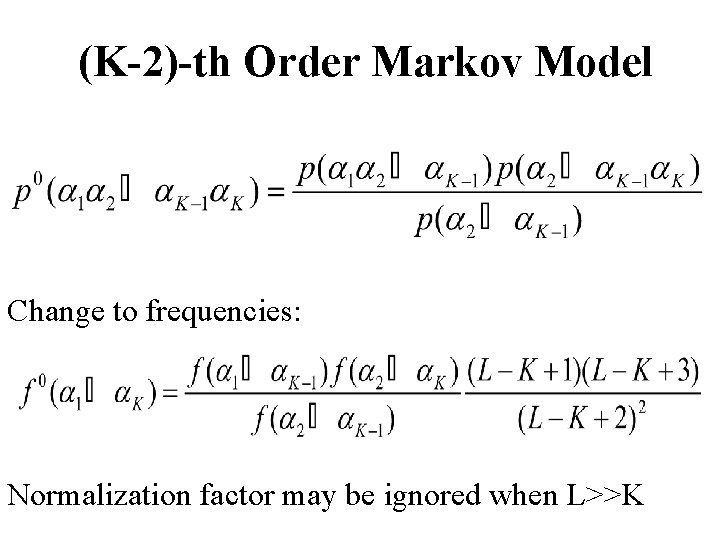
(K-2)-th Order Markov Model Change to frequencies: Normalization factor may be ignored when L>>K
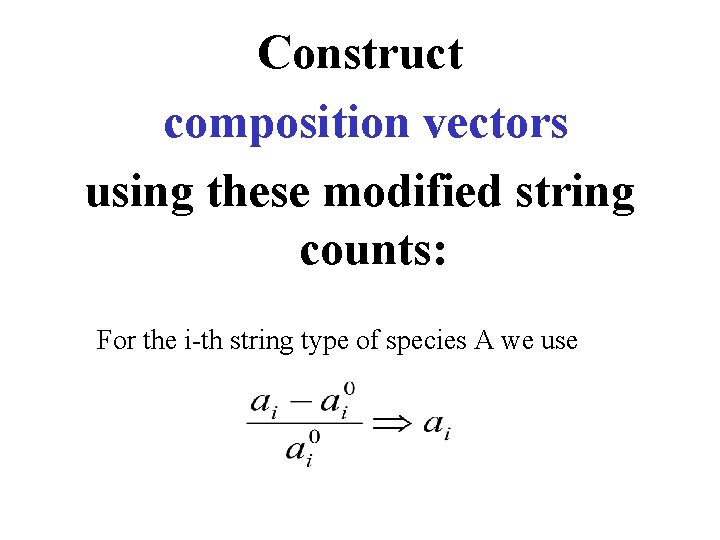
Construct composition vectors using these modified string counts: For the i-th string type of species A we use
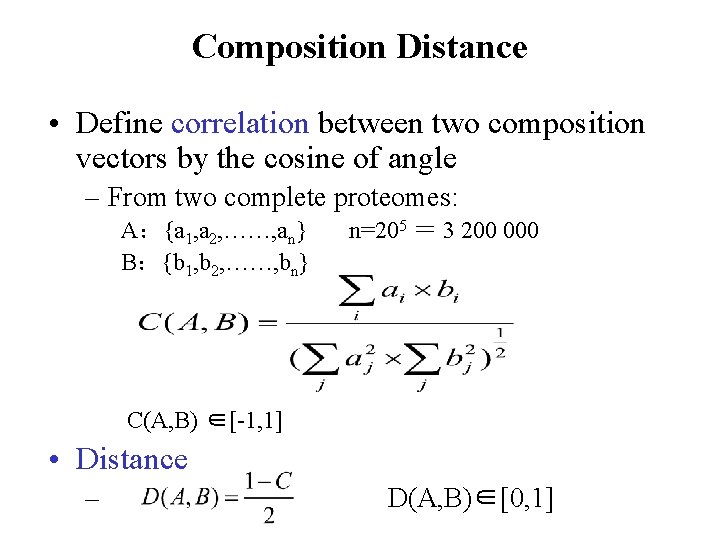
Composition Distance • Define correlation between two composition vectors by the cosine of angle – From two complete proteomes: A:{a 1, a 2, ……, an} B:{b 1, b 2, ……, bn} n=205 = 3 200 000 C(A, B) ∈[-1, 1] • Distance – D(A, B)∈[0, 1]
Protein Class vs. Whole Proteome • Trees based on collection of ribosomal proteins (SSU + LSU): ribosomal proteins are interwoven with r. RNA to form functioning complex; results consistent with SSU r. RNA trees • Trees based on collection of aminoacyl-t. RNA synthetases (AARS). Trees based on single AARS were not good. Trees based on all 20 AARSs taken together much better but not as good as that based on r. Proteins.
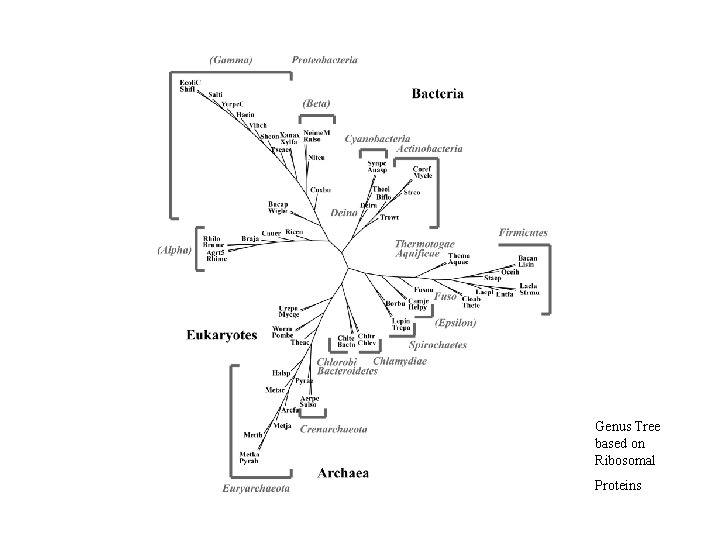
Genus Tree based on Ribosomal Proteins
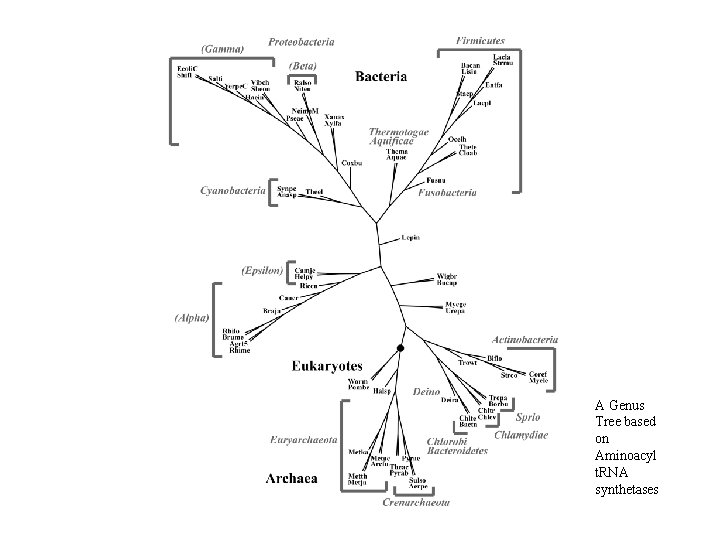
A Genus Tree based on Aminoacyl t. RNA synthetases
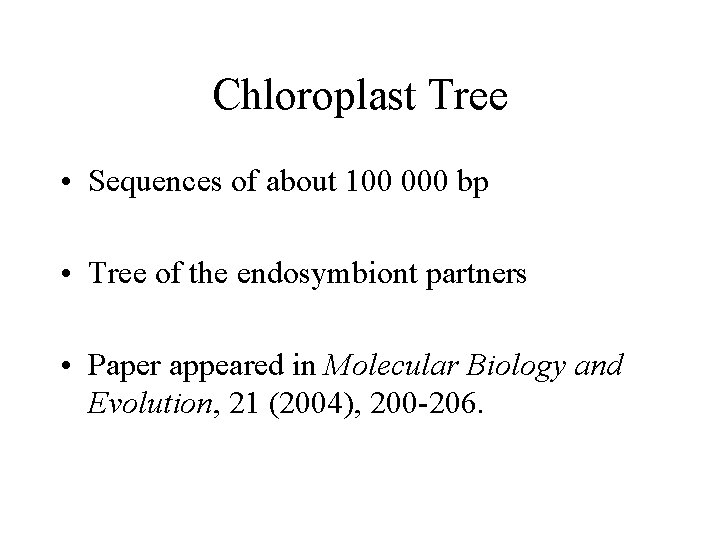
Chloroplast Tree • Sequences of about 100 000 bp • Tree of the endosymbiont partners • Paper appeared in Molecular Biology and Evolution, 21 (2004), 200 -206.
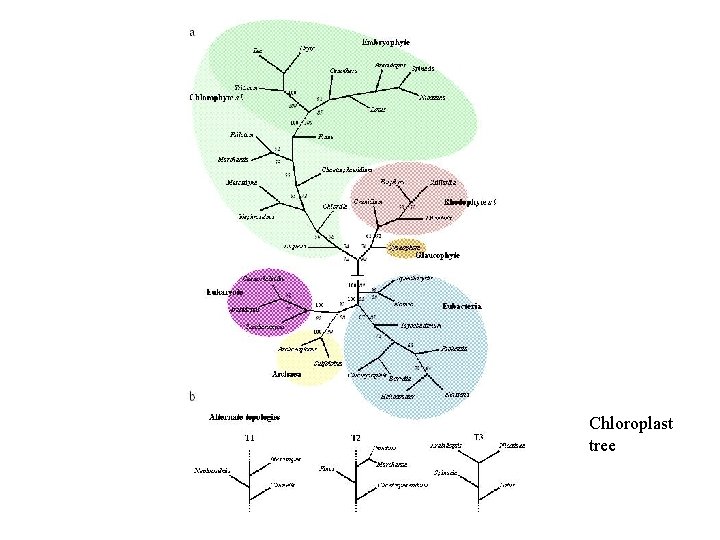
Chloroplast tree

Coronaviruses including Human SARS-Co. V • Sequences of tens kilo bases • SARS squence: about 29730 bases • Paper published in Chinese Science Bulletin, 48(12), 1170 -1174 (26 June 2003)
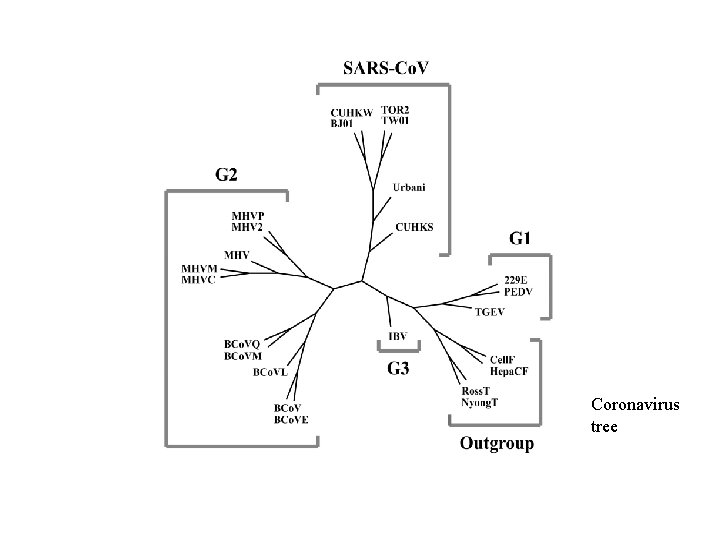
Coronavirus tree
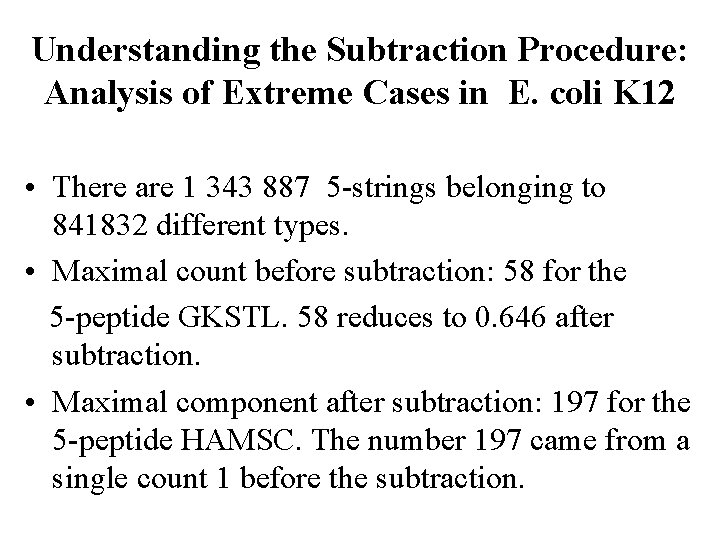
Understanding the Subtraction Procedure: Analysis of Extreme Cases in E. coli K 12 • There are 1 343 887 5 -strings belonging to 841832 different types. • Maximal count before subtraction: 58 for the 5 -peptide GKSTL. 58 reduces to 0. 646 after subtraction. • Maximal component after subtraction: 197 for the 5 -peptide HAMSC. The number 197 came from a single count 1 before the subtraction.
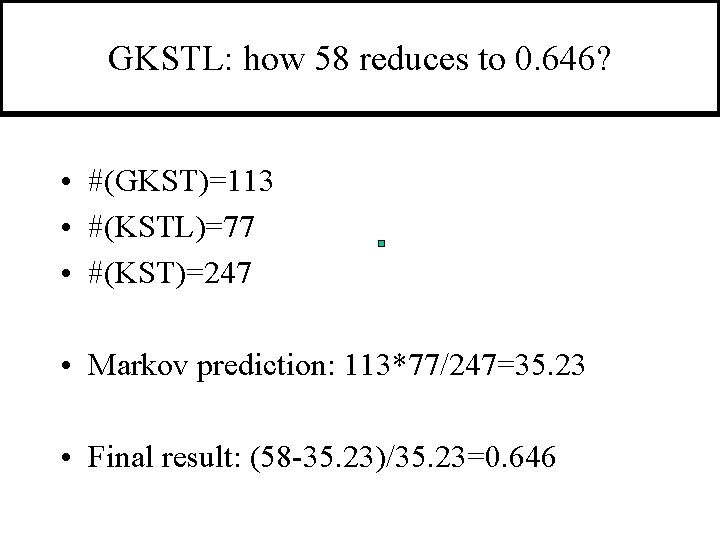
GKSTL: how 58 reduces to 0. 646? • #(GKST)=113 • #(KSTL)=77 • #(KST)=247 • Markov prediction: 113*77/247=35. 23 • Final result: (58 -35. 23)/35. 23=0. 646

HAMSC: how 1 grows to 197? • #(HAMS)=1 • #(AMSC)=1 • #(AMS)=198 • Markov prediction: 1*1/198=1/198 • Final result: (1 -1/198)/(1/198)=197
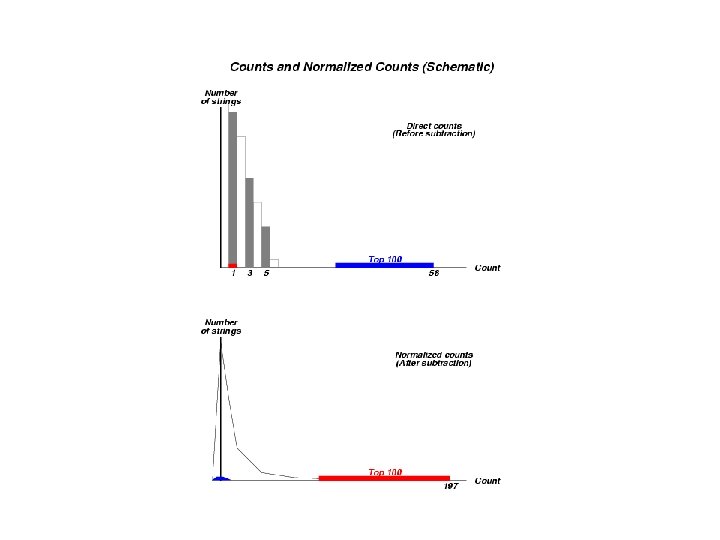

6121 Exact Matches of GKSTL In PIR Rel. 1. 26 with >1. 2 Mil Proteins • These 6121 matches came from a diverse taxonomic assortment from virus to bacteria to fungi to plants and animals including human being • In the parlance of classic cladistics GKSTL contributes to plesiomorphic characters that should be eliminated in a strict phylogeny • The subtraction procedure did the job.
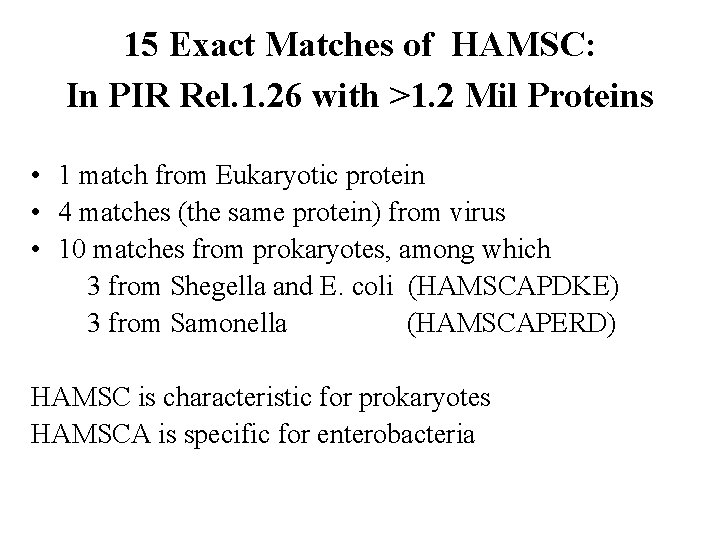
15 Exact Matches of HAMSC: In PIR Rel. 1. 26 with >1. 2 Mil Proteins • 1 match from Eukaryotic protein • 4 matches (the same protein) from virus • 10 matches from prokaryotes, among which 3 from Shegella and E. coli (HAMSCAPDKE) 3 from Samonella (HAMSCAPERD) HAMSC is characteristic for prokaryotes HAMSCA is specific for enterobacteria
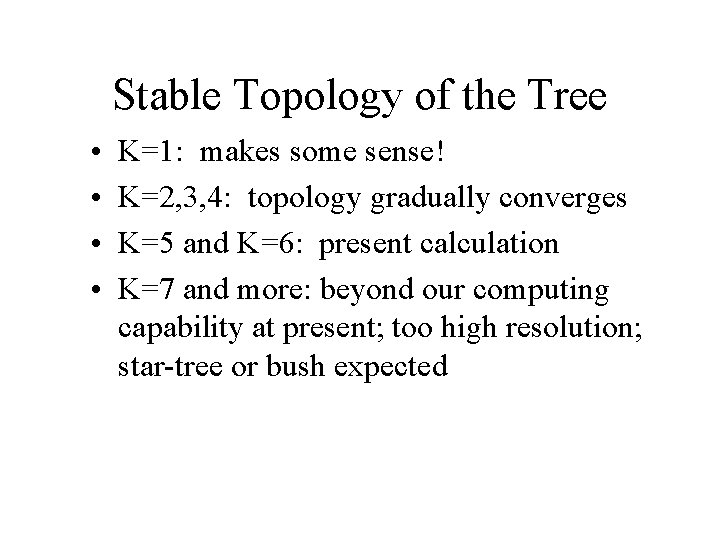
Stable Topology of the Tree • • K=1: makes some sense! K=2, 3, 4: topology gradually converges K=5 and K=6: present calculation K=7 and more: beyond our computing capability at present; too high resolution; star-tree or bush expected

Statistical Test of the Tree • Bootstrap versus Jack knife • Bootstrap in sequence alignments • “Bootstrap” by random selections from the AA-sequence pool • A time consuming job • 180 bootstraps for 72 species
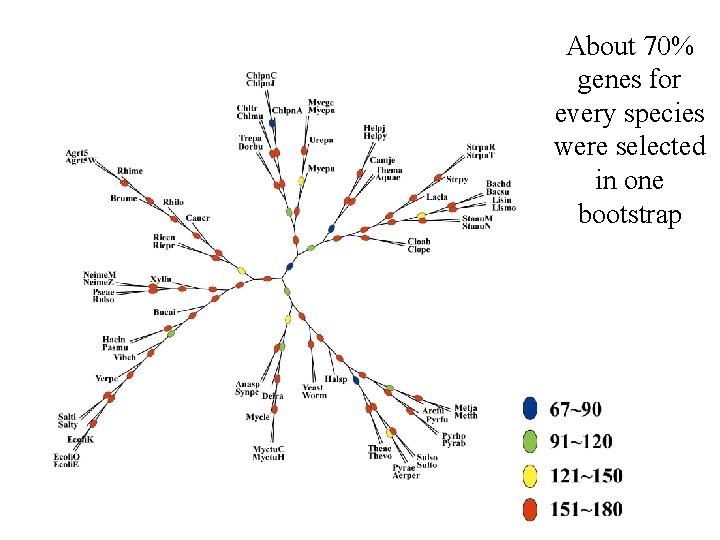
About 70% genes for every species were selected in one bootstrap
“K-string Picture” of Evolution • K=5 ->3 200 000 points in space of 5 -strings • K=6 ->64 000 points • In the primordial soup: short polypeptides of a limited assortment • Evolution by growth, fusion, mutation leads to diffusion in the string space • String space not saturated yet

The Problem of Higher Taxa • 1974: Bacteria as a separate kingdom • 1994: Archaea and Bacetria as two domains • The relation of higher taxa? Much debate among bacteriologists; but some hints from our trees and other whole-genome trees • No wonder: taxonomists of all walks disagree on grouping and palcing higher taxa

References • J Qi, B Wang, BL Hao, J. Mol. Evol. 58 (2004) 1 -11. (109=16 A + 87 B + 6 E) • KH Chu, J Qi, ZG Yu, V Ahn, Mol. Biol. Evol. 21(2004) 200 -206. (Chloroplasts) • L Gao, HB Wei, J Qi, YG Sun, BL Hao, Chinese Sci. Bull. 48(2003) 1170 -1174. (Coronavirus, SARSCo. V) • HB Wei, J Qi, BL Hao, Science in China, 34(2) (2004) 186 -199. (Using ribosomal and aminoacyl t. RNA synthetases) • BL Hao, J Qi, J. Bioinf. & Comput. Biol. 2 (2004) 1 -19. (A review with 132=16 A + 110 B + 6 E)
Summary § As composition vectors do not depend on genome size and gene content. The use of whole genome data is straightforward § Data independent on that of 16 S r. RNA § Method different from that based on SSU r. RNA § Results agree with SSU r. RNA trees and the Bergey’s Manual § Hint on groupings of higher taxa § A method without “free parameters”: data in, tree out § Possibility of an automatic and objective classification tool for prokaryotes

Conclusion: The phylogeny has met taxonomy. The Tree of Life is saved! There is phylogenetic information in the prokaryotic proteomes. Time to work on molecular definition of taxa. Thank you!

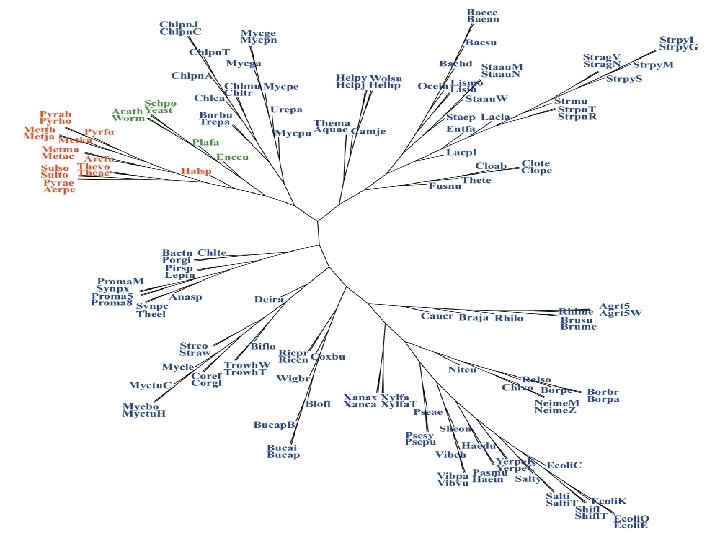
A Protein Tree for 154 Organisms From 88 Genera (K=5) 17 Archaea (12 genera, 17 species) 131 Bacteria (70 genera, 105 species) 6 Eukaryotes

- Slides: 46