Whole Genome of Ancient Human is Decoded Gene













- Slides: 51

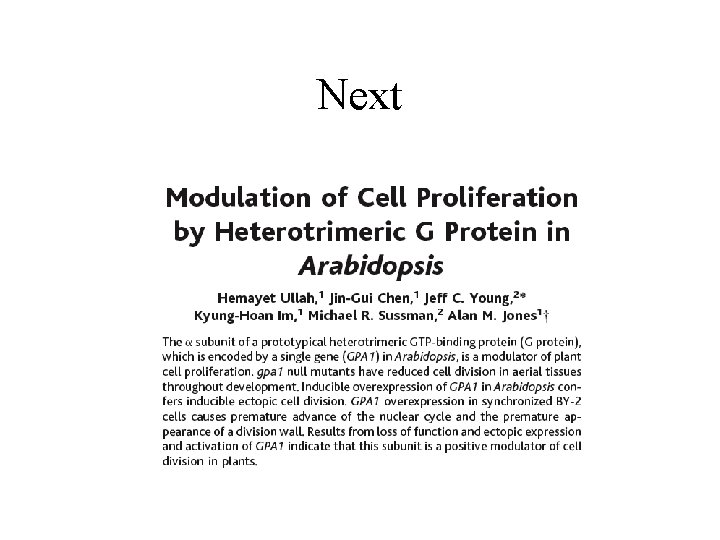
Whole Genome of Ancient Human is Decoded

Gene Expression new frontiers …the processes by which information contained in genes and genomes is decoded by cells, in order to produce molecules that determine the phenotypes observed in organisms, – transcription is controlled so that the correct DNA sequences are expressed as m. RNA in the right cells, at the right time, and in the right amount. - and, now we’re learning - processing and translation of m. RNA is further controlled (through RNA/Protein complexes), via ancient, conserved processes.

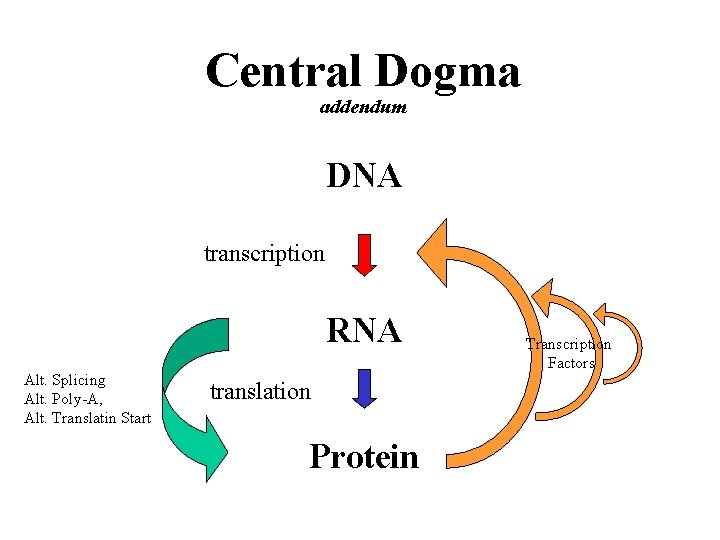
Central Dogma addendum DNA transcription RNA Alt. Splicing Alt. Poly-A, Alt. Translatin Start translation Protein Transcription Factors

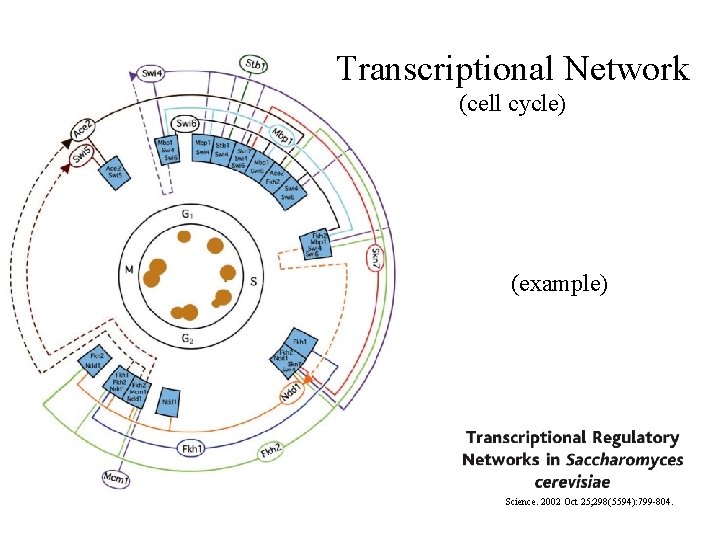
Transcriptional Network (cell cycle) (example) Science. 2002 Oct 25; 298(5594): 799 -804.

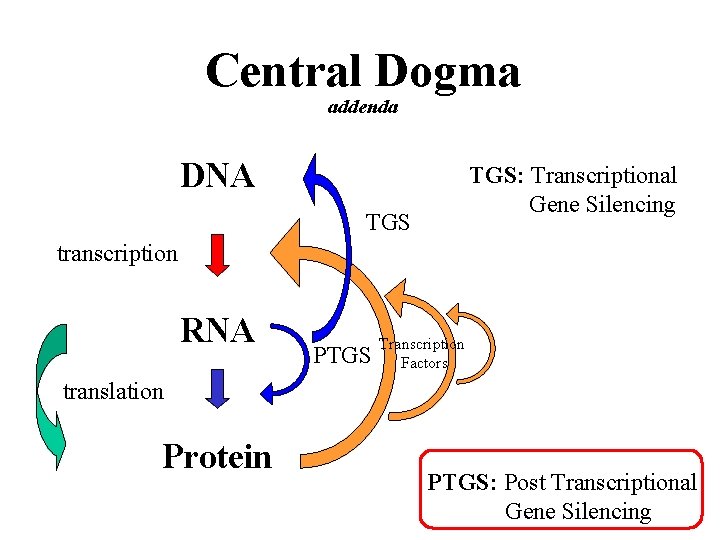
Central Dogma addenda DNA TGS: Transcriptional Gene Silencing transcription RNA PTGS Transcription Factors translation Protein PTGS: Post Transcriptional Gene Silencing

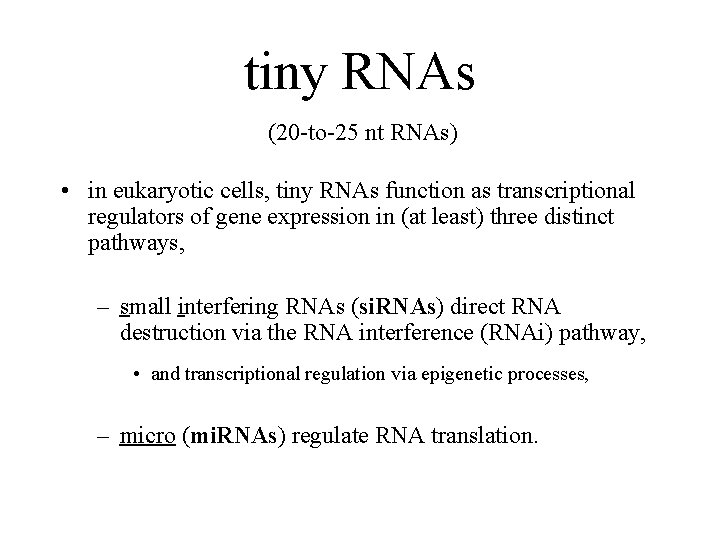
tiny RNAs (20 -to-25 nt RNAs) • in eukaryotic cells, tiny RNAs function as transcriptional regulators of gene expression in (at least) three distinct pathways, – small interfering RNAs (si. RNAs) direct RNA destruction via the RNA interference (RNAi) pathway, • and transcriptional regulation via epigenetic processes, – micro (mi. RNAs) regulate RNA translation.

Ancient History (1) Cell 75, 843 (1993) Some development timing genes code for short anti-sense molecules, …appeared to be unique to C. elegans.

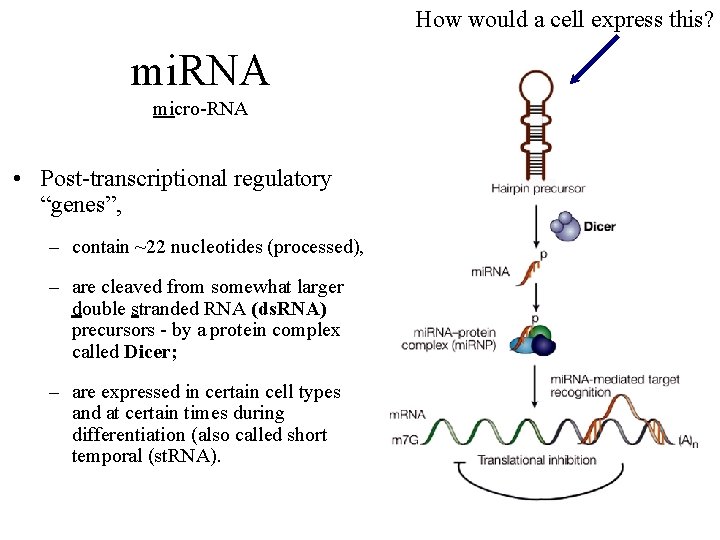
How would a cell express this? mi. RNA micro-RNA • Post-transcriptional regulatory “genes”, – contain ~22 nucleotides (processed), – are cleaved from somewhat larger double stranded RNA (ds. RNA) precursors - by a protein complex called Dicer; – are expressed in certain cell types and at certain times during differentiation (also called short temporal (st. RNA).

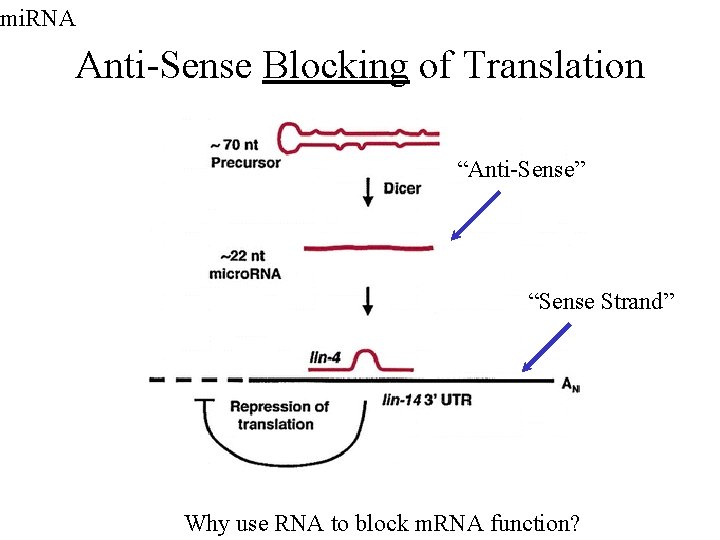
mi. RNA Anti-Sense Blocking of Translation “Anti-Sense” “Sense Strand” Why use RNA to block m. RNA function?

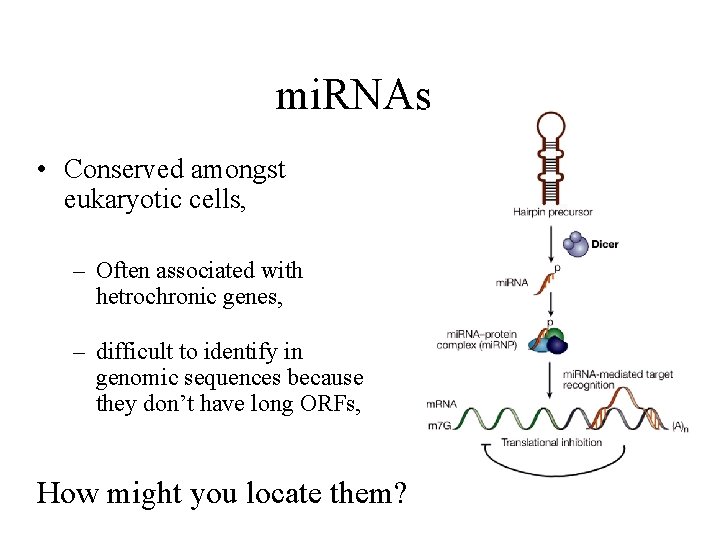
mi. RNAs • Conserved amongst eukaryotic cells, – Often associated with hetrochronic genes, – difficult to identify in genomic sequences because they don’t have long ORFs, How might you locate them?

Ancient History (II) (co-suppression) Transgene expression often decreases as the copy number of transgenes increased.

Over Expression Studies • Make a gene construct with, – Structural Gene, – Active promoter (often from a virus promoter), – Marker gene to be able to determine transformation. Marker Gene (w/P) Active promoter Gene of Interest • Expect, – Higher levels of protein, – Gene-dosage phenotypes, – Glorious publication. Frequent Results: no protein produced, scorn from senior scientists.

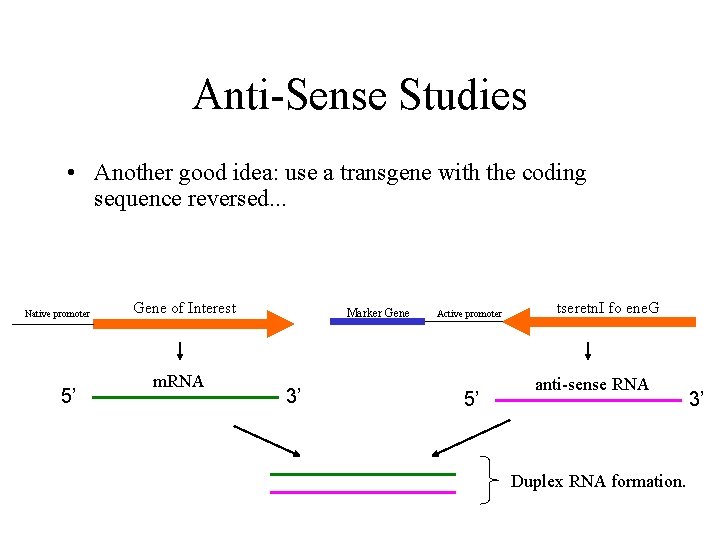
Anti-Sense Studies • Another good idea: use a transgene with the coding sequence reversed. . . Native promoter 5’ Gene of Interest m. RNA Marker Gene 3’ Active promoter 5’ tseretn. I fo ene. G anti-sense RNA Duplex RNA formation. 3’

Expected Results • Low, to no detectable single stranded transcript, • Low, to no protein products, • Glorious publication detailing gene function. Actual Results (Wacky) • Phenotypes ranged from death to over-expression, • Transcript levels were also extremely variable, • Scorn from senior scientists.

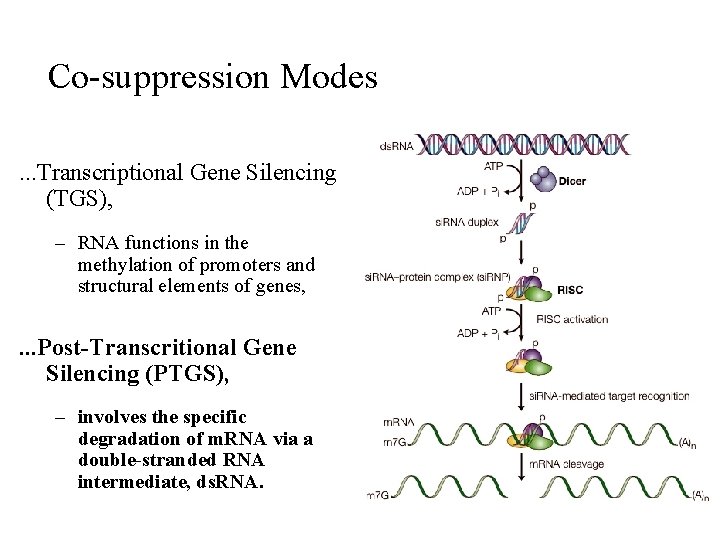
Co-suppression Modes. . . Transcriptional Gene Silencing (TGS), – RNA functions in the methylation of promoters and structural elements of genes, . . . Post-Transcritional Gene Silencing (PTGS), – involves the specific degradation of m. RNA via a double-stranded RNA intermediate, ds. RNA.

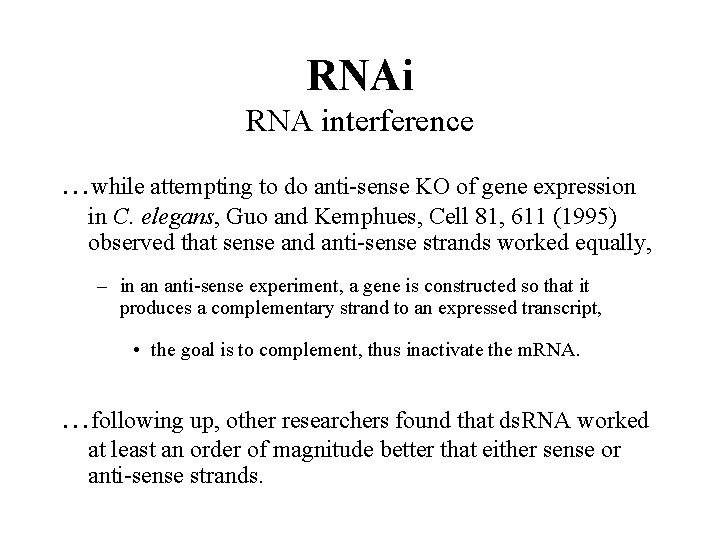
RNAi RNA interference . . . while attempting to do anti-sense KO of gene expression in C. elegans, Guo and Kemphues, Cell 81, 611 (1995) observed that sense and anti-sense strands worked equally, – in an anti-sense experiment, a gene is constructed so that it produces a complementary strand to an expressed transcript, • the goal is to complement, thus inactivate the m. RNA. . following up, other researchers found that ds. RNA worked at least an order of magnitude better that either sense or anti-sense strands.

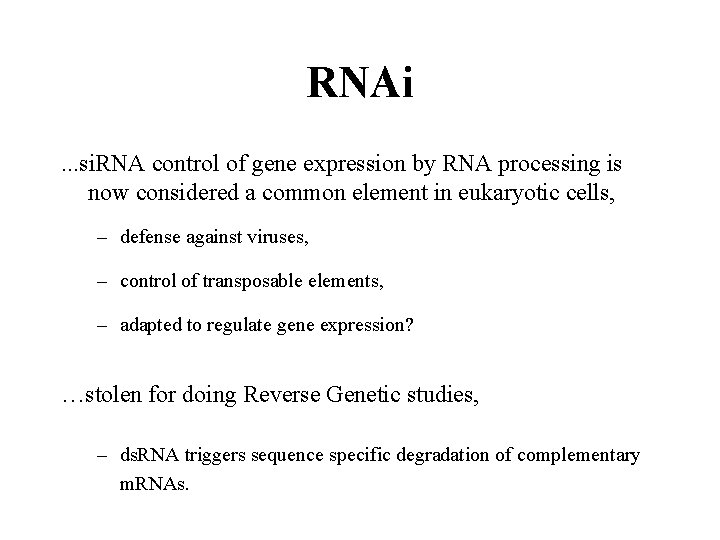
RNAi. . . si. RNA control of gene expression by RNA processing is now considered a common element in eukaryotic cells, – defense against viruses, – control of transposable elements, – adapted to regulate gene expression? …stolen for doing Reverse Genetic studies, – ds. RNA triggers sequence specific degradation of complementary m. RNAs.

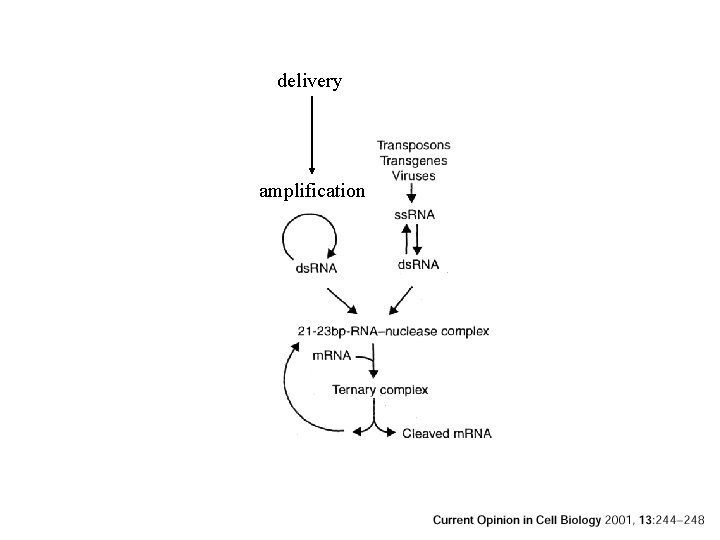
delivery amplification

Today Nature 408: 331 - 336

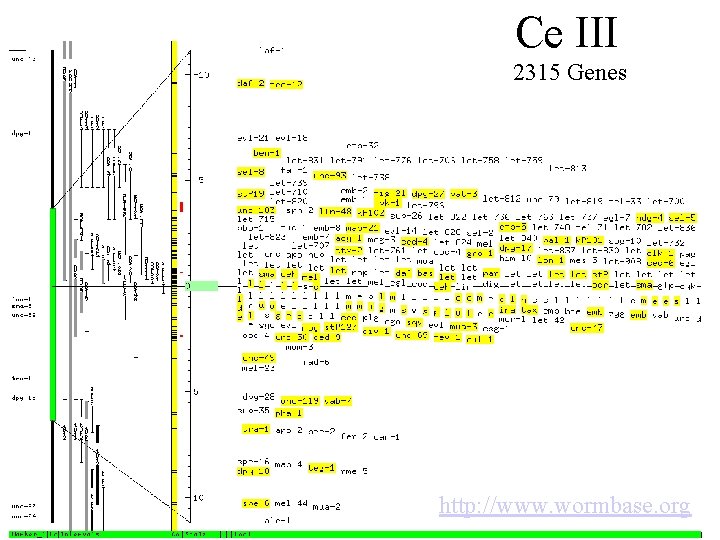
Ce III 2315 Genes http: //www. wormbase. org

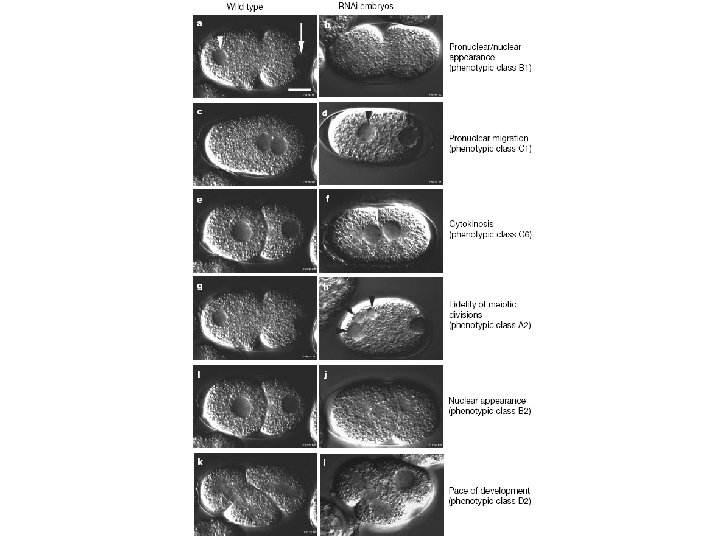
Functional Genomics The Question(s) Can we establish a high throughput system to assign cellular function to genes identified in metazoans? - using cell division and associated processes as the scorable phenotype, In the process, can we learn about… – – cell division genes, embryology, general development, anything else?

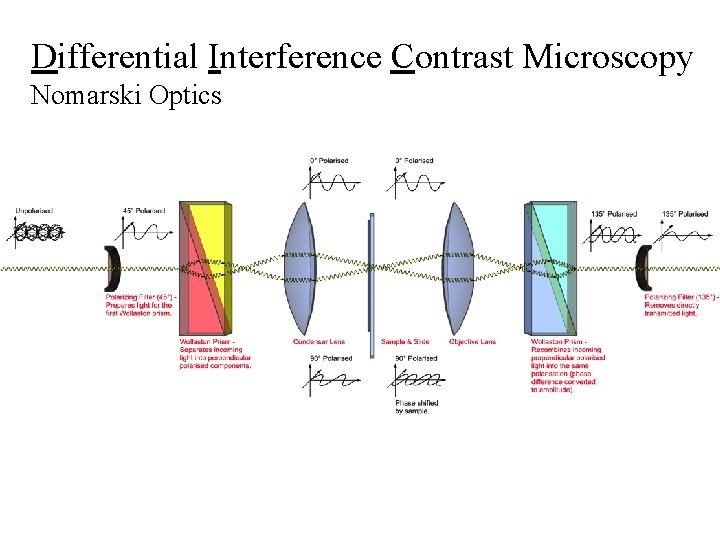
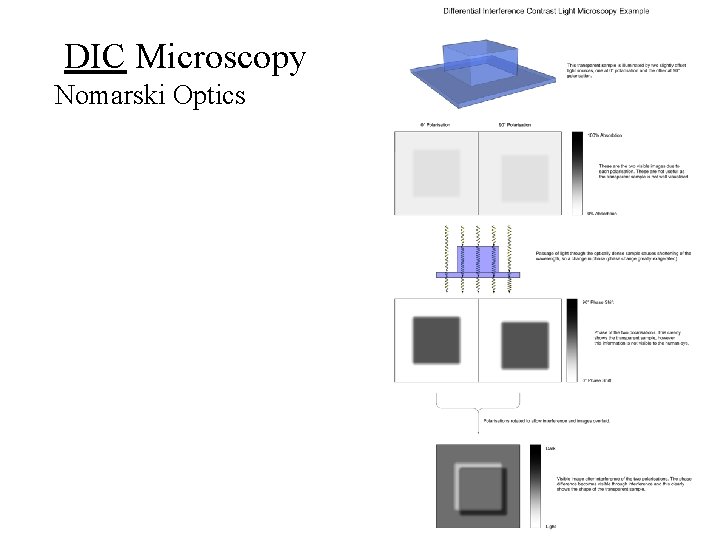
Differential Interference Contrast Microscopy Nomarski Optics

DIC Microscopy Nomarski Optics

Reverse Genetics Knockomics, Knockology. . . Sequence to Phenotype to Function

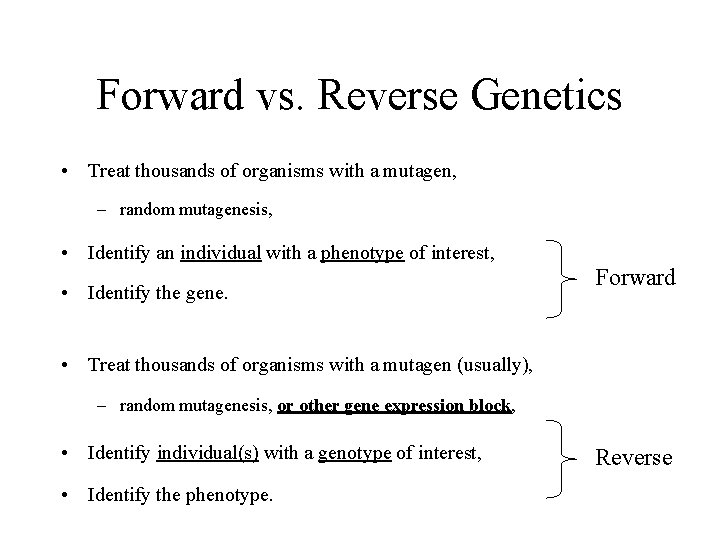
Forward vs. Reverse Genetics • Treat thousands of organisms with a mutagen, – random mutagenesis, • Identify an individual with a phenotype of interest, • Identify the gene. Forward • Treat thousands of organisms with a mutagen (usually), – random mutagenesis, or other gene expression block, • Identify individual(s) with a genotype of interest, • Identify the phenotype. Reverse

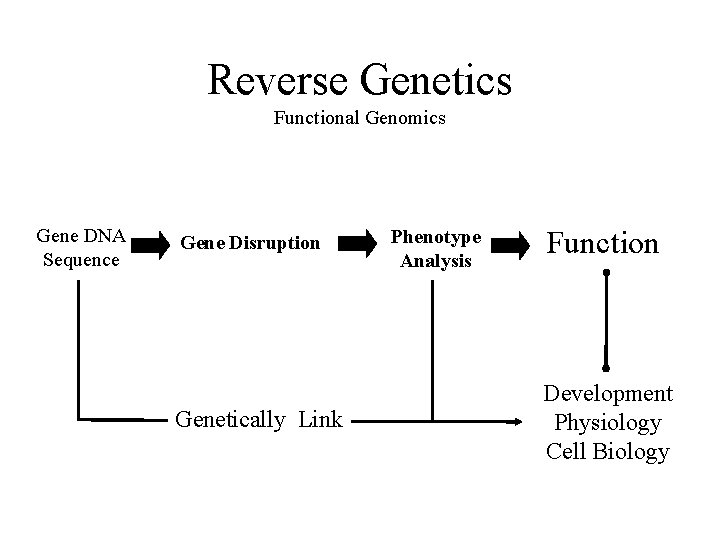
Reverse Genetics Functional Genomics Gene DNA Sequence Gene Disruption Genetically Link Phenotype Analysis Function Development Physiology Cell Biology

New Data, New Technology new paradigms • The C. elegans genome is sequenced, and we can identify 2315 candidate sequences on Chromosome III. • We can see cell division through a microscope, and further, we are able to identify many abnormalities. • We have RNAi technology at hand to selectively “knock down” any gene we are interested in. Further, RNAi can be added to cells prior to fertilization, mitosis commences after fertilization.

Reverse Genetics Discovery Research (High Throughput) • Few, if any, hypothesis going in, • High throughput, (2232 genes), • Lots of “negative” results, (87. 1% of the genes tested), • Value is in (12. 9%)… – the analysis of the data in concert with annotations in the data sets and references in the literature, – the generation of materials for further “hypothesis” - or - “discovery” driven research.

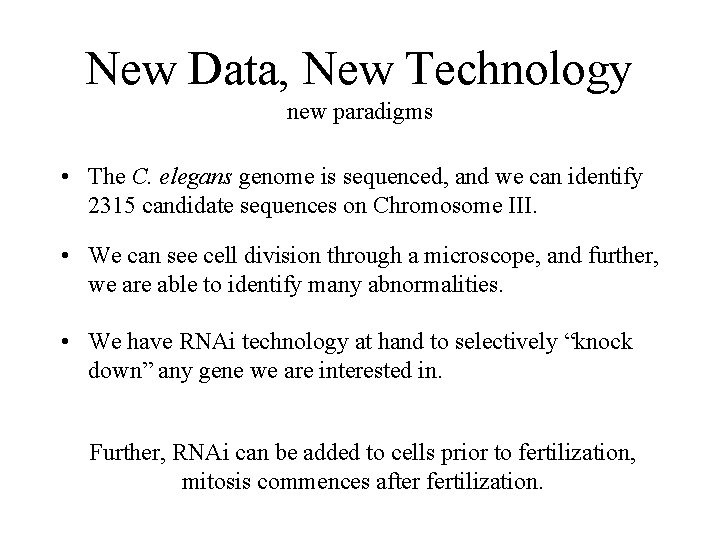
ds. RNAs (I) Where do they come from? • PCR primer pairs were designed for each of the genes discovered via bioinformatic analysis of the sequenced chromosome, – and confirmed through EST sequences, or experimental expression studies, – shortest region > 500 bp, or > 90% of ORF. gene ds. DNA

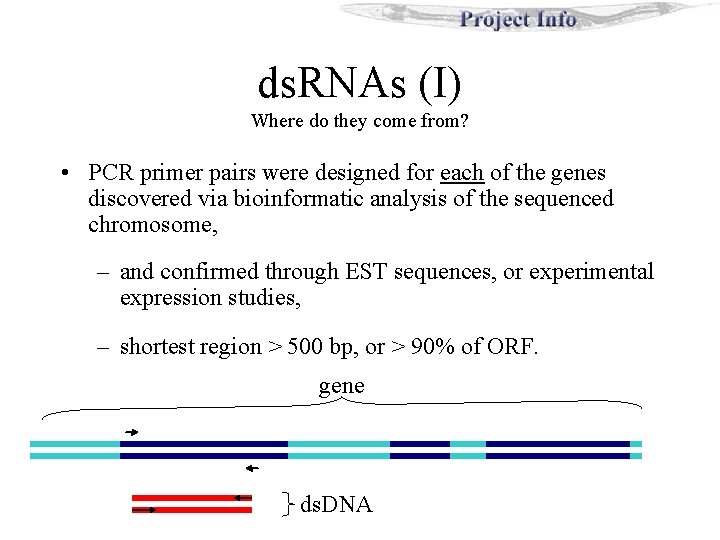
ds. RNAs (II) PCR Primers + • T 3 or T 7 promoter sequences were included in the PCR primers. . . T 3 sequence tacked onto the reverse primer. gene 5’ - GTAATACGACTCACTATAGGG GCTAAGCTATTCGATGCTA - 3’ T 7 promoter sequence gene specific sequence

T 3 and T 7 RNA Polymerase • Bacteriophage T 3 and T 7 RNA polymerases are DNAdependent RNA polymerases with high sequence specificity for T 3 or T 7 promoters. • T 3 and T 7 RNA polymerases synthesize RNA 5' to 3'. • These enzymes are isolated from an overproducing recombinant E. coli clone, and are available commercially.

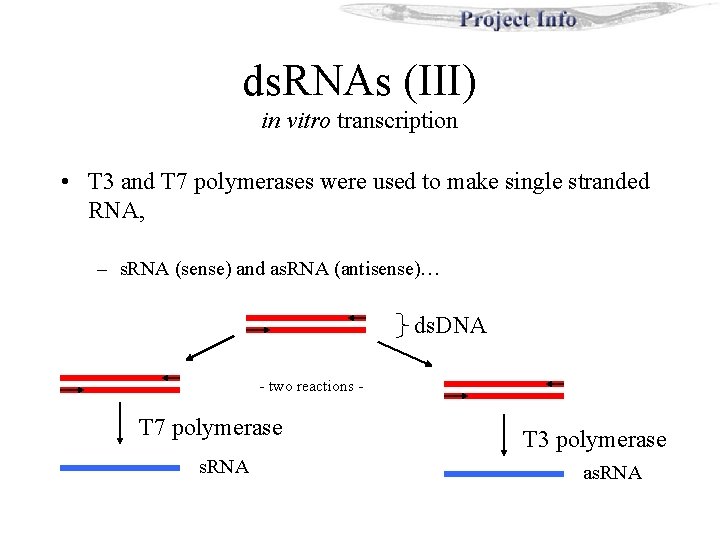
ds. RNAs (III) in vitro transcription • T 3 and T 7 polymerases were used to make single stranded RNA, – s. RNA (sense) and as. RNA (antisense)… ds. DNA - two reactions - T 7 polymerase s. RNA T 3 polymerase as. RNA

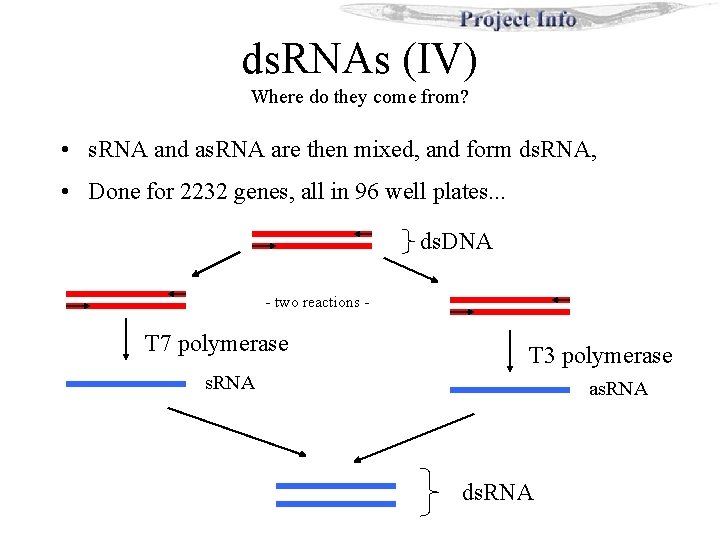
ds. RNAs (IV) Where do they come from? • s. RNA and as. RNA are then mixed, and form ds. RNA, • Done for 2232 genes, all in 96 well plates. . . ds. DNA - two reactions - T 7 polymerase T 3 polymerase s. RNA as. RNA ds. RNA

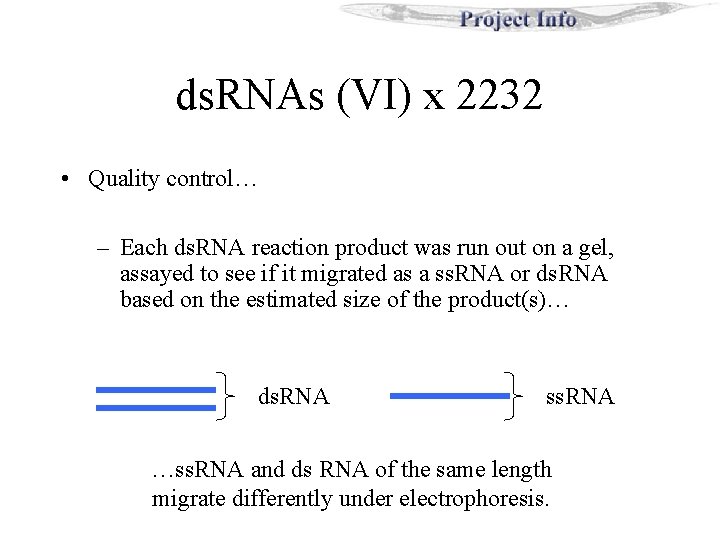
ds. RNAs (VI) x 2232 • Quality control… – Each ds. RNA reaction product was run out on a gel, assayed to see if it migrated as a ss. RNA or ds. RNA based on the estimated size of the product(s)… ds. RNA ss. RNA …ss. RNA and ds RNA of the same length migrate differently under electrophoresis.

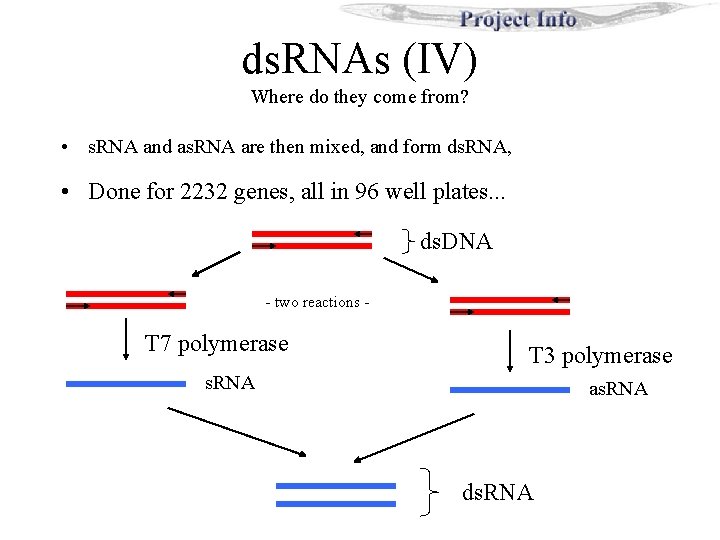
ds. RNAs (IV) Where do they come from? • s. RNA and as. RNA are then mixed, and form ds. RNA, • Done for 2232 genes, all in 96 well plates. . . ds. DNA - two reactions - T 7 polymerase T 3 polymerase s. RNA as. RNA ds. RNA

Then What? • ds. RNAs (was) injected. . . into the gonads of adult wildtype hermaphrodites, which were left at 20 °C for 24 h, • Embryos were then removed analyzed for potential defects in cell-division processes, capturing 1 image every 5 s using time-lapse Nomarski Differential Interference Contrast (DIC) microscopy, • A minimum of three embryos from three different worms were filmed from shortly after fertilization until the fourcell stage. http: //fire. biol. wwu. edu/young/470/rnai_movies. html

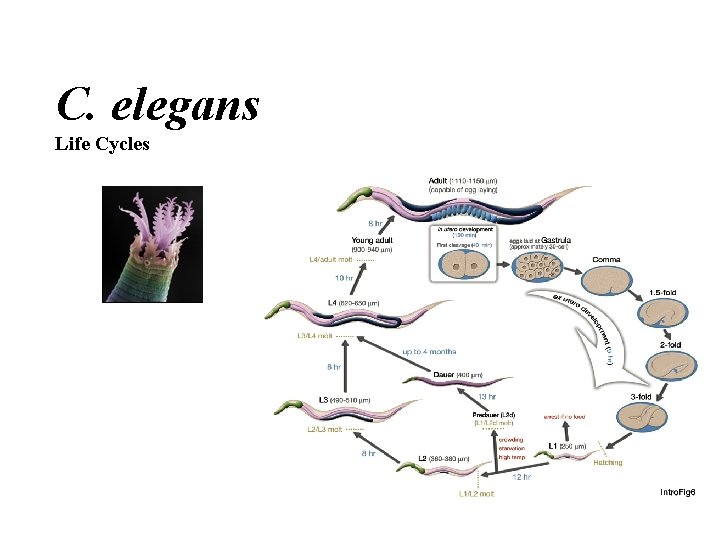
C. elegans Life Cycles

And More… Progeny Tests • Three animals were transferred to a fresh plate 24 h after injection, and left at 20 °C. – Two days later, the plate was inspected with a stereomicroscope (20– 40 x magnification) for the presence of eggs, F 1 larvae and their developmental stage (normally L 2–L 4). – Two days after that, the plate was inspected for the presence of F 1 adults (normally >100), their overall body morphology and the presence of F 2 progeny. • Partially penetrant embryonic lethality and subtle developmental defects were not scored in this analysis. • ds. RNAs that gave rise to defects in less than 5% of the adult progeny were not considered as being associated with a phenotype.

But? • It’s supposed to be high throughput, so experiments were designed to minimize the time required, – in part to make the acquisition of so much “meaningless” data palatable (89. 1%), – in part because it is a whole lot of work no matter how you approach it, • Remember, along with discovery, this experiment was designed to establish a workable paradigm for future large scale analysis of metazoan (and other complex) organisms.

So, First establish reliability • Injected 13 ds. RNAs targeted to known components of the cell division process, – all 13 known mutations were observable using DIC photography, • This control tested RNAi efficiency, and the efficacy of DIC phenotype scoring. . . 13 of 13 genes were disrupted, based on clear DIC image acquisition.

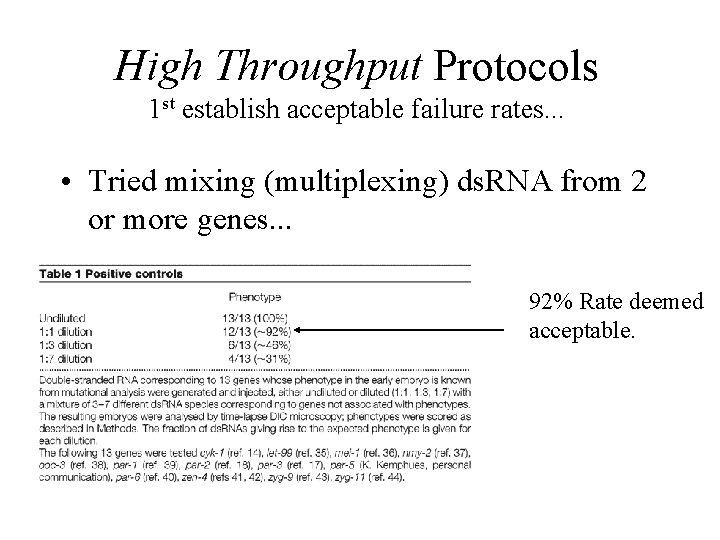
High Throughput Protocols 1 st establish acceptable failure rates. . . • Tried mixing (multiplexing) ds. RNA from 2 or more genes. . . 92% Rate deemed acceptable.

1. Then did it, 2. Then checked the results. . . • When a phenotype was observed… – to see which of the two ds. RNAs caused the phenotype, fresh worms were injected with the ds. RNA (one at a time), – genomic sequence was examined to make sure that only the ds. RNA targeted gene was responsible, • Gene families, • Miscalled ORFs.

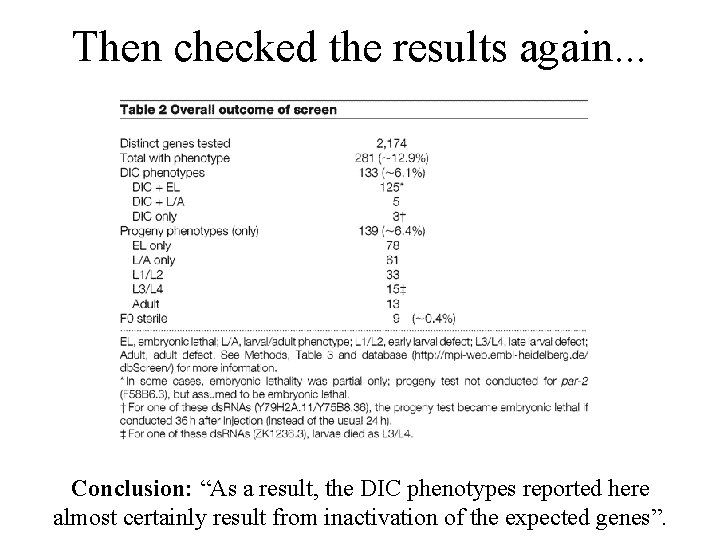
Then checked the results again. . . Conclusion: “As a result, the DIC phenotypes reported here almost certainly result from inactivation of the expected genes”.


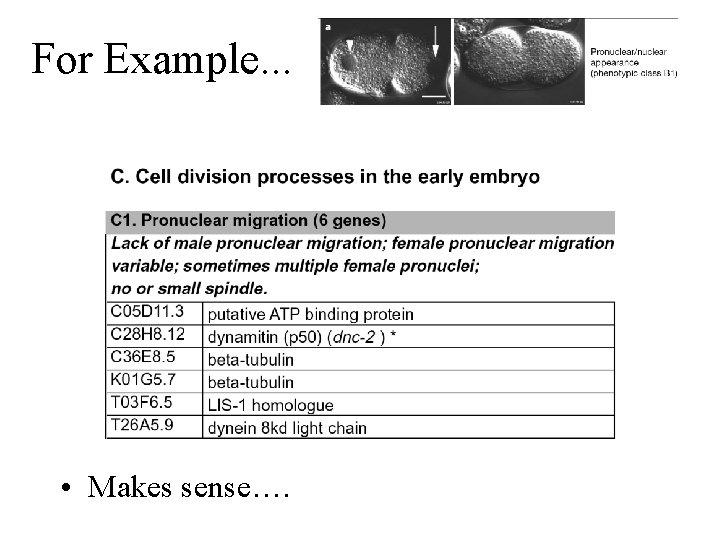
For Example. . . • Makes sense….

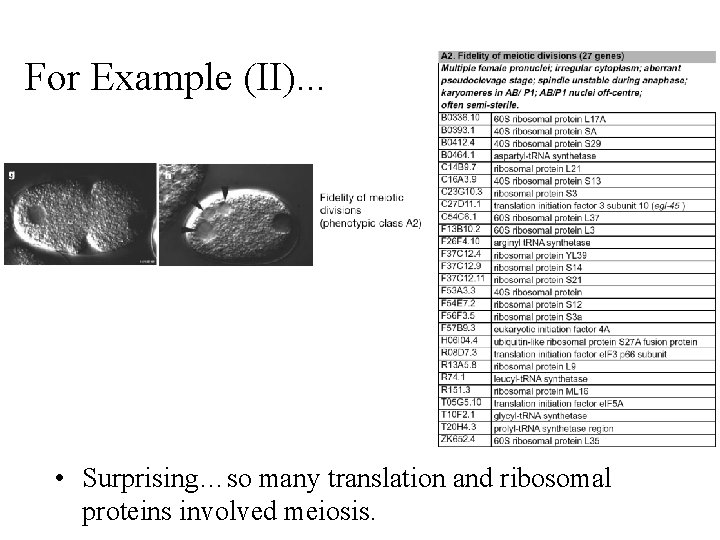
For Example (II). . . • Surprising…so many translation and ribosomal proteins involved meiosis.

Forward vs. Reverse Scorecard • 7 of 7 known chromosome III DIC observable, early embryo phenotypes observed, • 9 of 14 late embryo phenotypes observed, • 9 of 31 larvae/adult phenotypes observed. 7 of 7 known, plus 126 new genes!

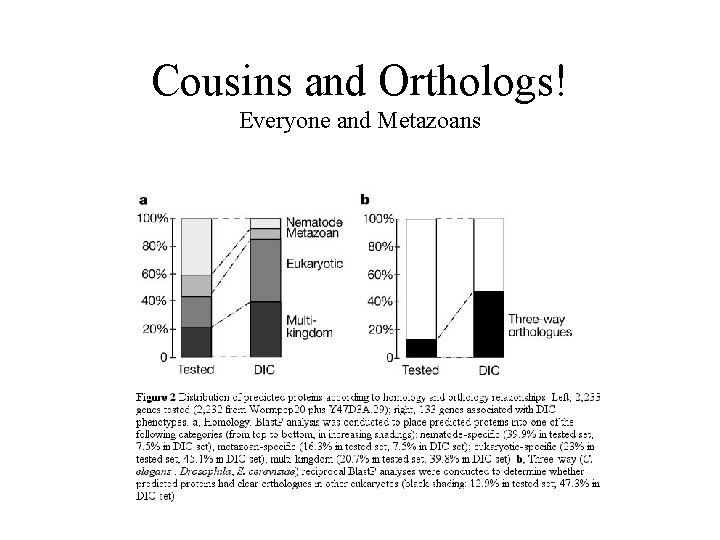
Cousins and Orthologs! Everyone and Metazoans

Successful? • High throughput: Yes, • Fidelity: Yes, 7/7, • Discovery: Yes, > 100 new genes involved in early embryo development, especially cell division, • Helpful to Metazoan biologists?

Next

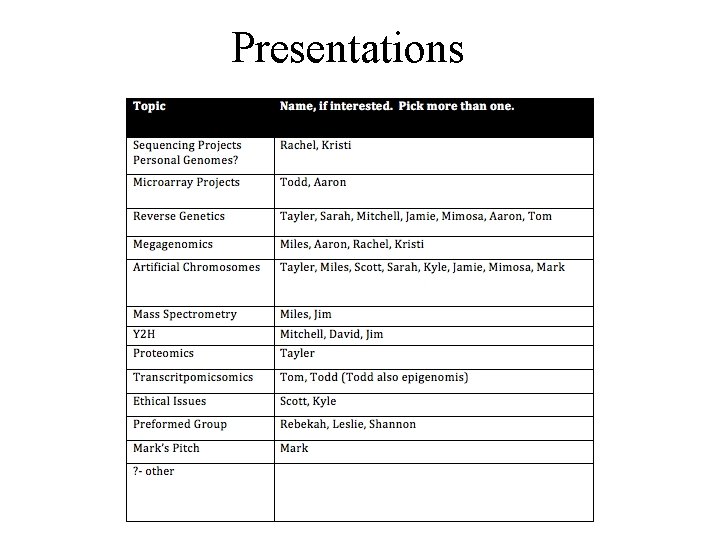
Presentations