What should a bioinformatician know about DNA sequencing

What should a bioinformatician know about DNA sequencing, and why?
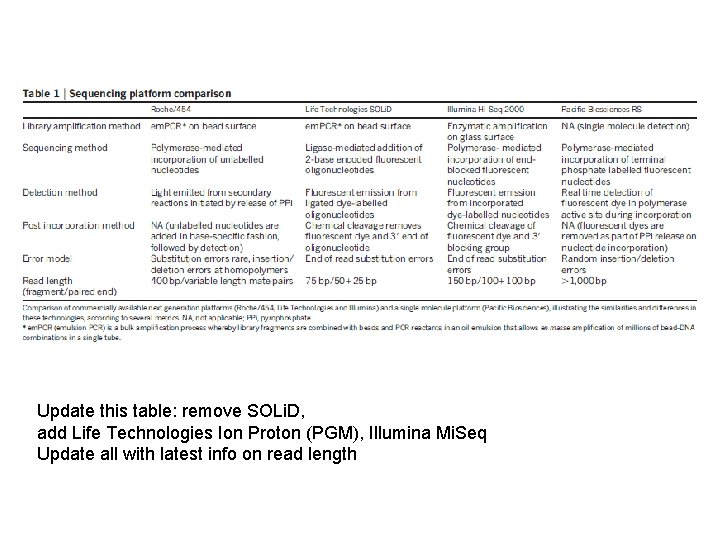
Update this table: remove SOLi. D, add Life Technologies Ion Proton (PGM), Illumina Mi. Seq Update all with latest info on read length

What are the error types and rates of the different platforms?
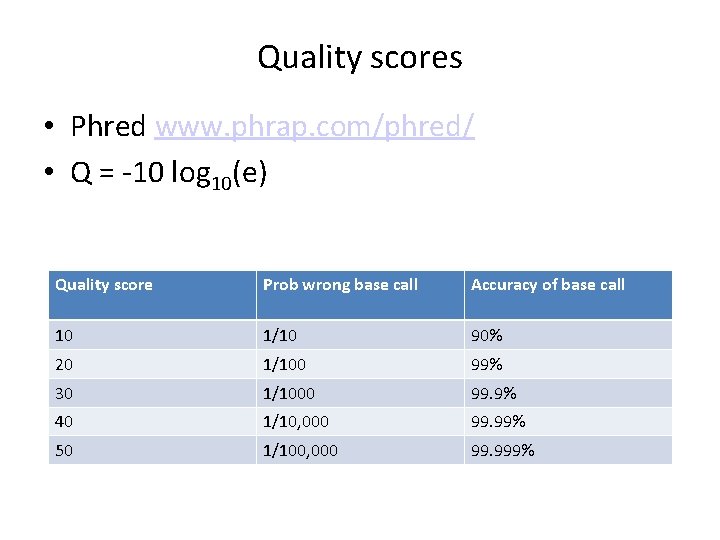
Quality scores • Phred www. phrap. com/phred/ • Q = -10 log 10(e) Quality score Prob wrong base call Accuracy of base call 10 1/10 90% 20 1/100 99% 30 1/1000 99. 9% 40 1/10, 000 99. 99% 50 1/100, 000 99. 999%
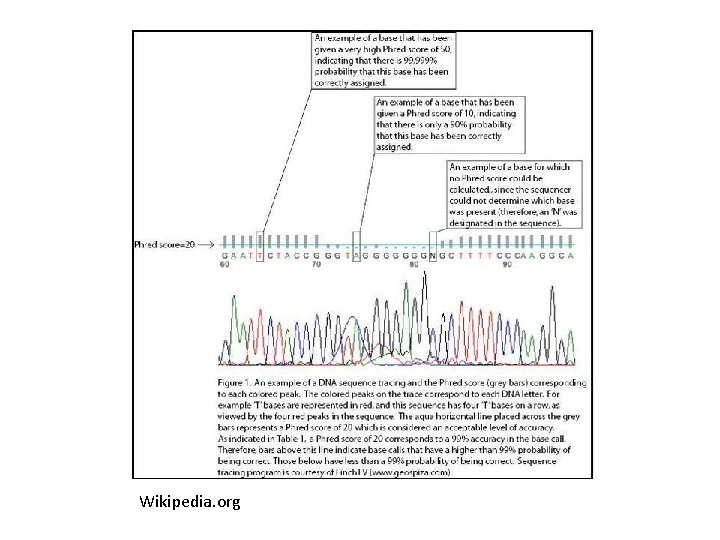
Wikipedia. org

FASTQ format 4 lines, sequence + quality scores @SEQ_ID (+optional description) GATTTGGGGTTCAAAGCAGTATCGATCAAATAGTAAATCCATTTGTTCAACTCACAGTTT + optional repeat of line 1, often left as just the + character to save space !''*((((***+))%%%++)(%%%%). 1***-+*''))**55 CCF>>>>>>CCCCCCC 65 But beware! At least 3 different FASTQ file standards, indistinguishable in format, but incompatible with each other Wikipedia. org
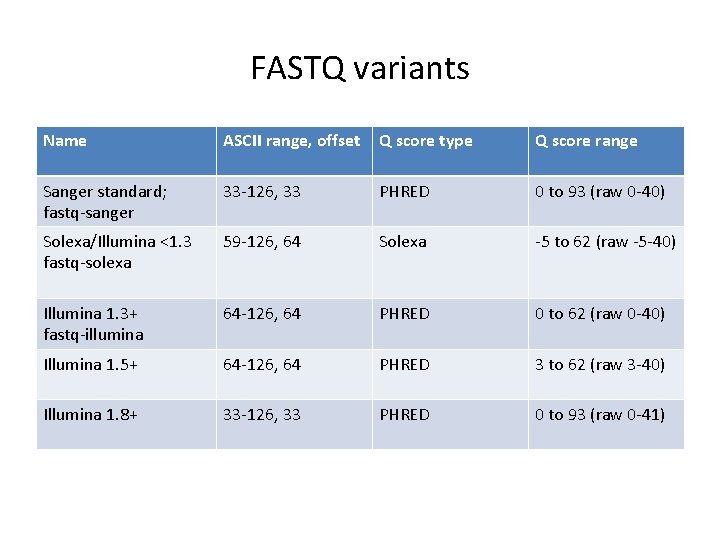
FASTQ variants Name ASCII range, offset Q score type Q score range Sanger standard; fastq-sanger 33 -126, 33 PHRED 0 to 93 (raw 0 -40) Solexa/Illumina <1. 3 fastq-solexa 59 -126, 64 Solexa -5 to 62 (raw -5 -40) Illumina 1. 3+ fastq-illumina 64 -126, 64 PHRED 0 to 62 (raw 0 -40) Illumina 1. 5+ 64 -126, 64 PHRED 3 to 62 (raw 3 -40) Illumina 1. 8+ 33 -126, 33 PHRED 0 to 93 (raw 0 -41)
What use is the quality score?

What factors should be considered in the choice of a DNA sequencing platform?
- Slides: 9