What evolutionary forces act on gene expression regulation



- Slides: 19

What evolutionary forces act on gene expression regulation? Before considering selection, it’s important to characterize how gene expression varies within and between species. What facilitates regulatory evolution? * Gene dispensibility Genes with variable expression within species are heavily enriched for non-essential genes * Genes with upstream TATA elements TATA regulation in yeast (and other organisms? ) is associated with variable expression * Redundancy Either gene or regulatory redundancy * Modularity in regulation Genes with more upstream elements or greater environmental responsiveness 1

What contributes to the evolution of gene expression? How many loci underlie expression variation? Few major effectors or many minor contributors? What are the mechanisms of expression evolution? Relative prominence of cis vs. trans effects? How much of expression variation has been selected for? 2

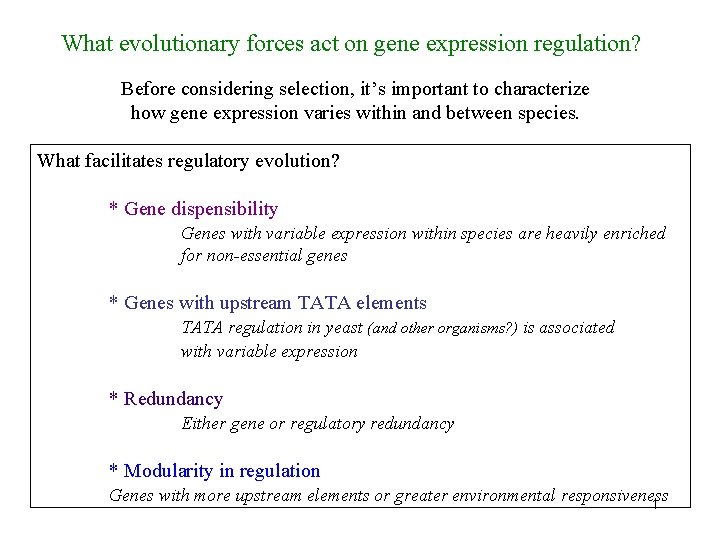
e. QTL mapping: understanding the genetic basis of expression variation first done in strains of yeast First done by Rachel Brem et al. 3 From Rockman & Kruglyak 2006

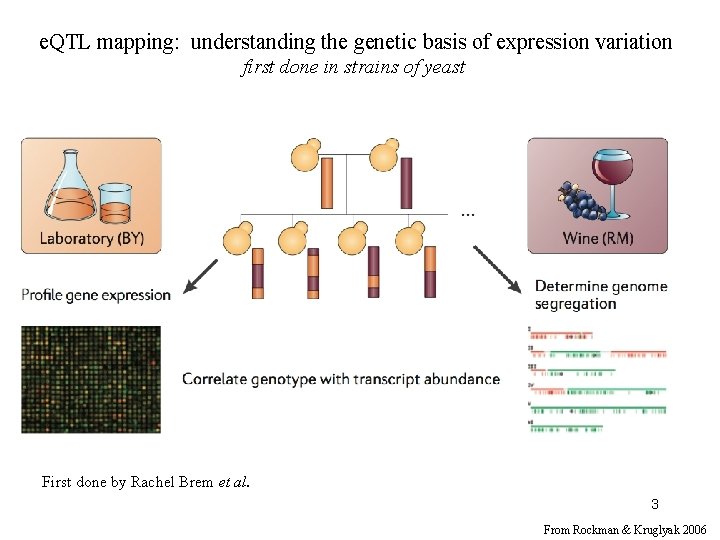
e. QTL mapping in yeast spore clones First done by Rachel Brem et al. 4 From Rockman & Kruglyak 2006

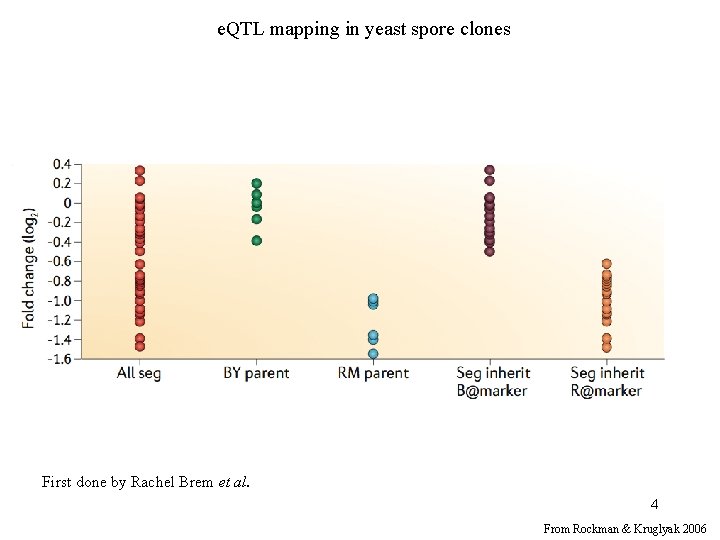
LOD threshold in standard QTL mapping (* 1 trait) • 1000 permutations 10% LOD score threshold: 3. 19 5% LOD score threshold: 3. 52 Challenge with e. QTL mapping is that there are thousands of traits. 5

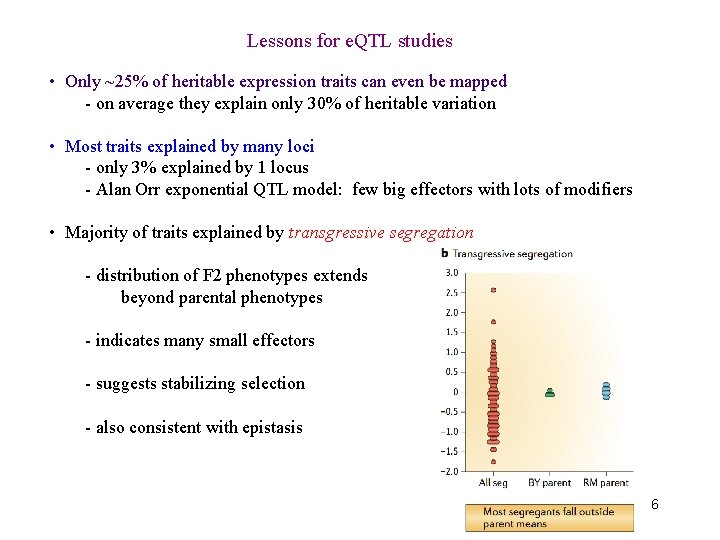
Lessons for e. QTL studies • Only ~25% of heritable expression traits can even be mapped - on average they explain only 30% of heritable variation • Most traits explained by many loci - only 3% explained by 1 locus - Alan Orr exponential QTL model: few big effectors with lots of modifiers • Majority of traits explained by transgressive segregation - distribution of F 2 phenotypes extends beyond parental phenotypes - indicates many small effectors - suggests stabilizing selection - also consistent with epistasis 6

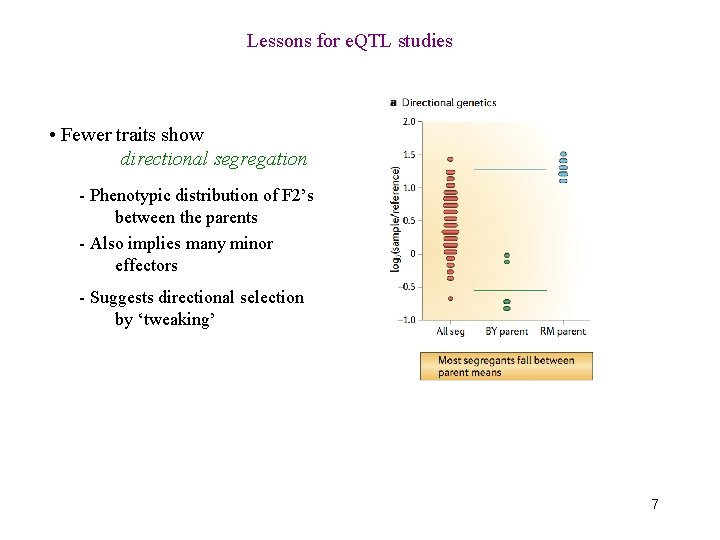
Lessons for e. QTL studies • Fewer traits show directional segregation - Phenotypic distribution of F 2’s between the parents - Also implies many minor effectors - Suggests directional selection by ‘tweaking’ 7

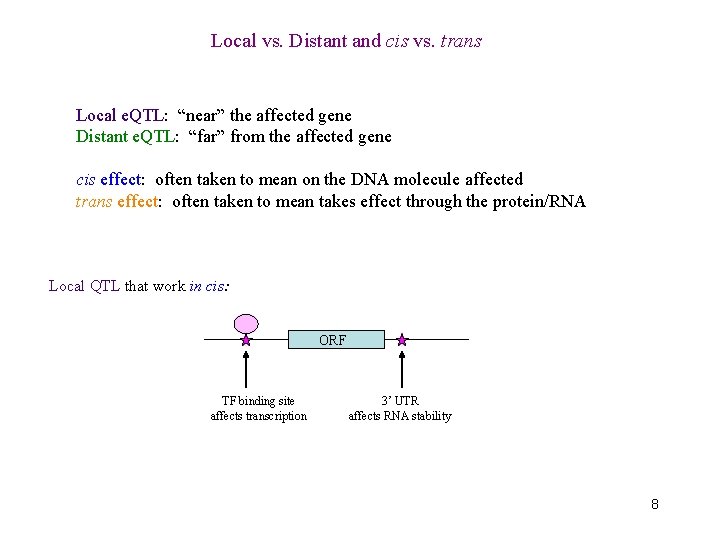
Local vs. Distant and cis vs. trans Local e. QTL: “near” the affected gene Distant e. QTL: “far” from the affected gene cis effect: often taken to mean on the DNA molecule affected trans effect: often taken to mean takes effect through the protein/RNA Local QTL that work in cis: ORF TF binding site affects transcription 3’ UTR affects RNA stability 8

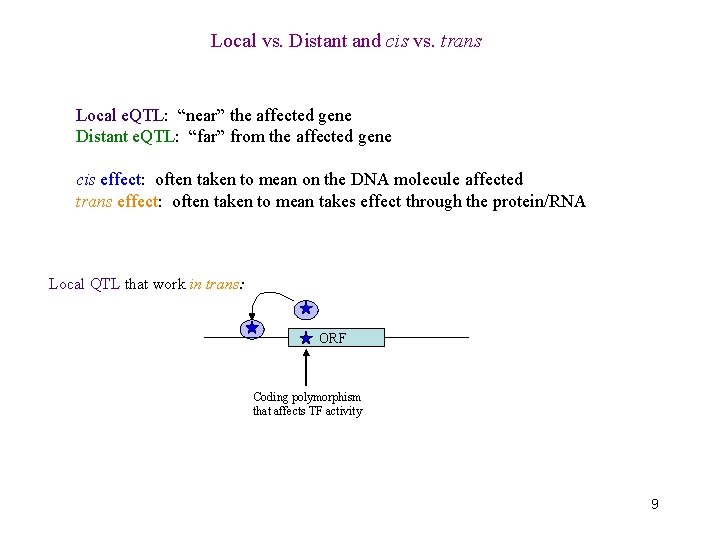
Local vs. Distant and cis vs. trans Local e. QTL: “near” the affected gene Distant e. QTL: “far” from the affected gene cis effect: often taken to mean on the DNA molecule affected trans effect: often taken to mean takes effect through the protein/RNA Local QTL that work in trans: ORF Coding polymorphism that affects TF activity 9

Local vs. Distant and cis vs. trans Local e. QTL: “near” the affected gene Distant e. QTL: “far” from the affected gene cis effect: often taken to mean on the DNA molecule affected trans effect: often taken to mean takes effect through the protein/RNA Distant QTL that work in trans: ORF 10

Local vs. Distant and cis vs. trans Local e. QTL: “near” the affected gene Distant e. QTL: “far” from the affected gene cis effect: often taken to mean on the DNA molecule affected trans effect: often taken to mean takes effect through the protein/RNA PHYSIOLOGY Distant QTL that work in trans: ORF Most trans acting effects are likely secondary responses (distantly-acting loci are NOT enriched for TFs) 11

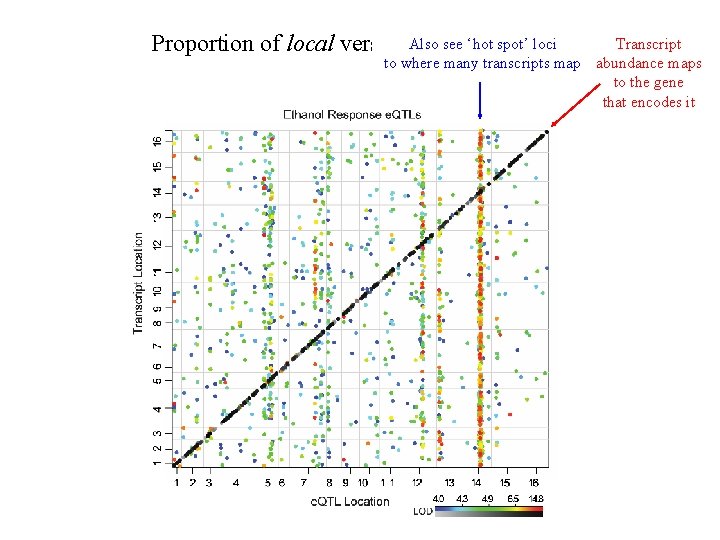
Also see ‘hot spot’ loci Proportion of local versus distant linkages Transcript to where many transcripts map abundance maps to the gene that encodes it

Local vs. Distant and cis vs. trans Which is more prevalent? Estimates vary: - Brem et al papers: ~25% traits explained by local polymorphs - other studies say close to 100% - Many MORE individual genes explained by distant polymorphs * but because many link to same loci, there are fewer distantly acting loci But … statistical challenges likely enrich for local polymorphisms: - FDR hurdle is higher for trans acting loci - cis (local) polymorphisms may have larger effect size - also depends on how “local” is defined 13

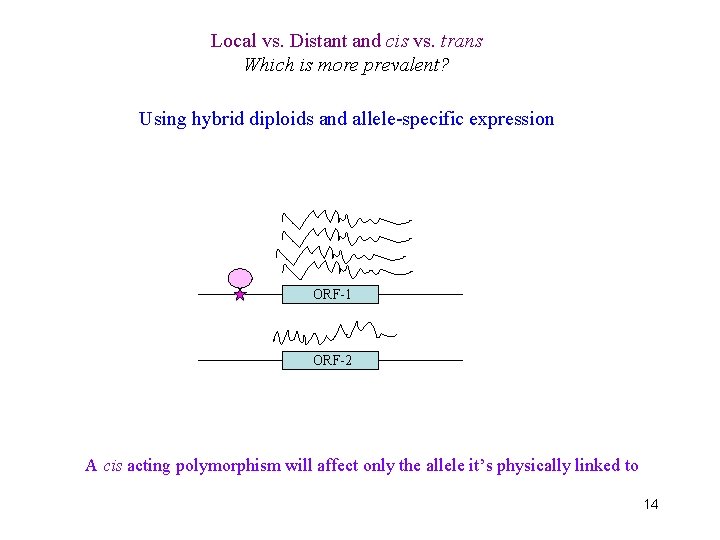
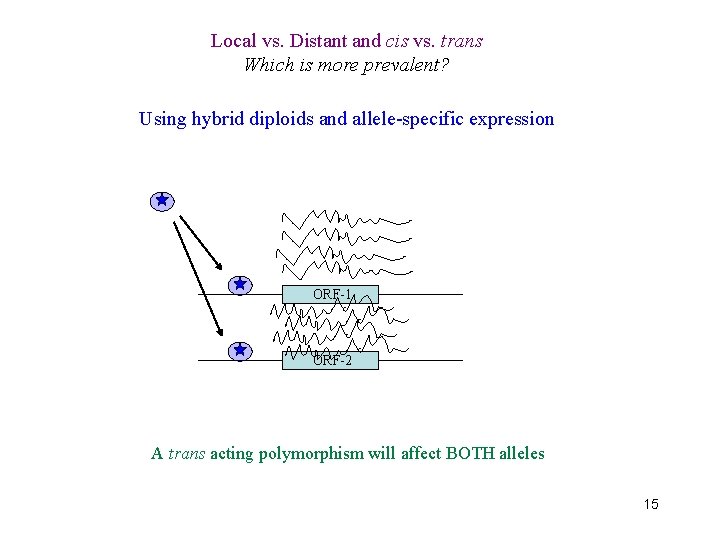
Local vs. Distant and cis vs. trans Which is more prevalent? Using hybrid diploids and allele-specific expression ORF-1 ORF-2 A cis acting polymorphism will affect only the allele it’s physically linked to 14

Local vs. Distant and cis vs. trans Which is more prevalent? Using hybrid diploids and allele-specific expression ORF-1 ORF-2 A trans acting polymorphism will affect BOTH alleles 15

Local vs. Distant and cis vs. trans Which is more prevalent? Tricia Wittkopp et al. 2004. Nature. - 29 differentially expressed genes between D. melanogaster & D. simulans: - Measured allele-specific expression in D. mel/D. sim hybrid with pyrosequencing 28 out of 29 show cis variation in expression 16 out of 29 affected by trans and cis variation Conclusion: cis-acting variation is more common to explain interspecific variation 16



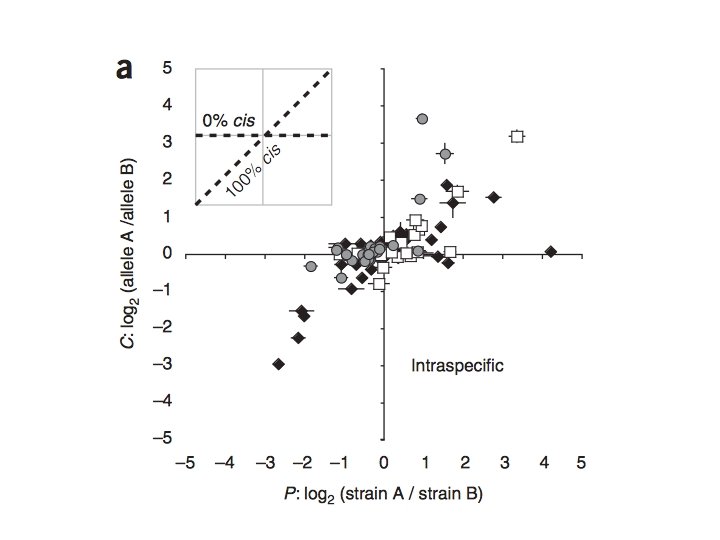
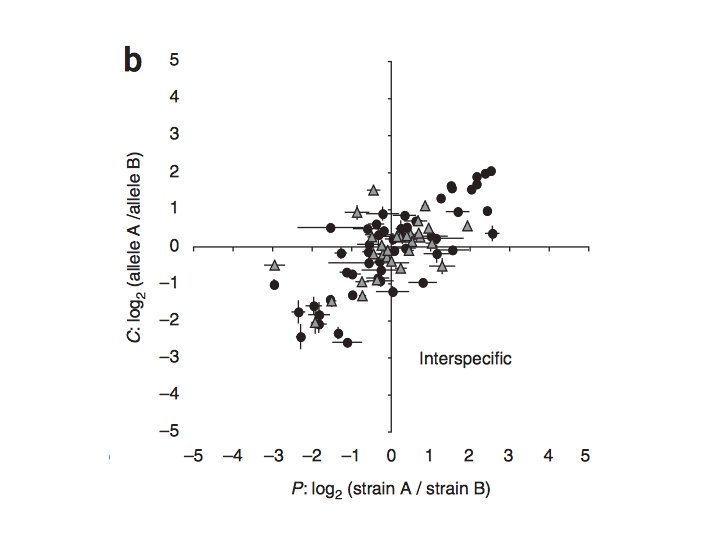
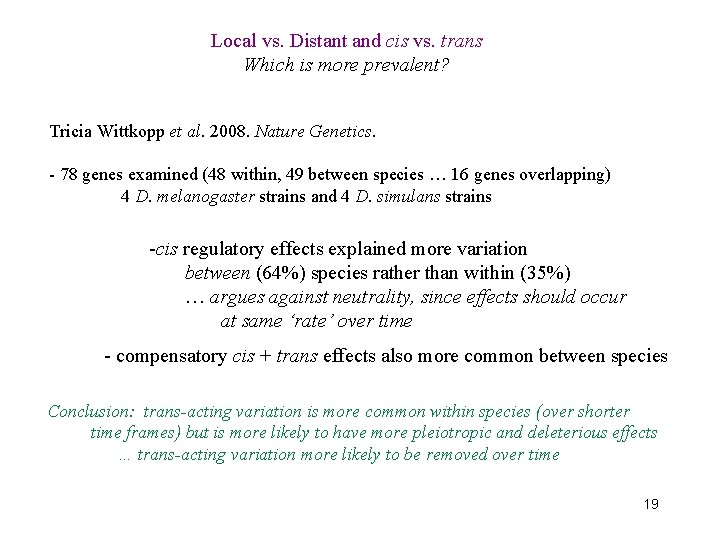
Local vs. Distant and cis vs. trans Which is more prevalent? Tricia Wittkopp et al. 2008. Nature Genetics. - 78 genes examined (48 within, 49 between species … 16 genes overlapping) 4 D. melanogaster strains and 4 D. simulans strains -cis regulatory effects explained more variation between (64%) species rather than within (35%) … argues against neutrality, since effects should occur at same ‘rate’ over time - compensatory cis + trans effects also more common between species Conclusion: trans-acting variation is more common within species (over shorter time frames) but is more likely to have more pleiotropic and deleterious effects … trans-acting variation more likely to be removed over time 19