What are the Patterns Of Nucleotide Substitution Within
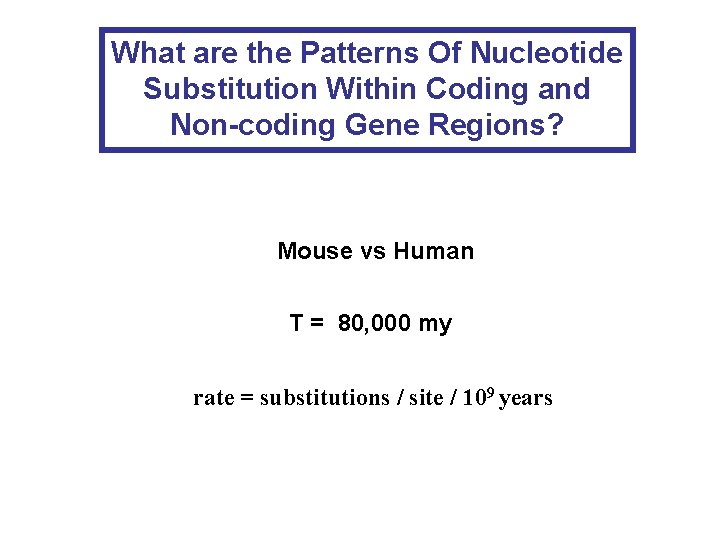
What are the Patterns Of Nucleotide Substitution Within Coding and Non-coding Gene Regions? Mouse vs Human T = 80, 000 my rate = substitutions / site / 109 years
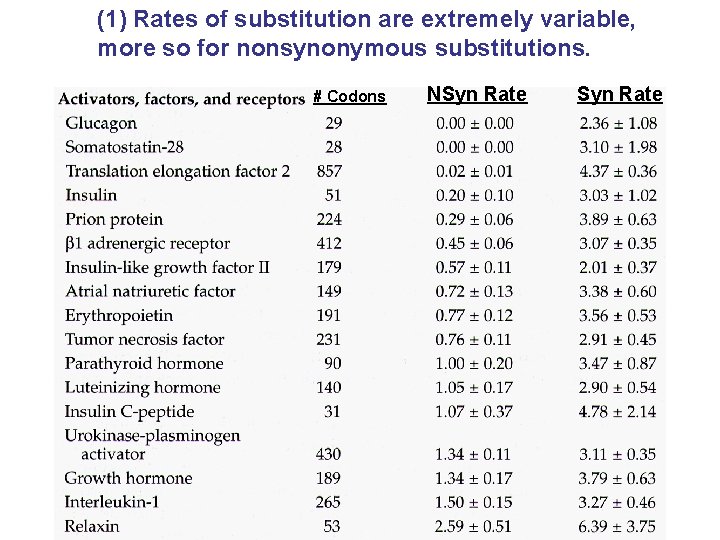
(1) Rates of substitution are extremely variable, more so for nonsynonymous substitutions. # Codons NSyn Rate
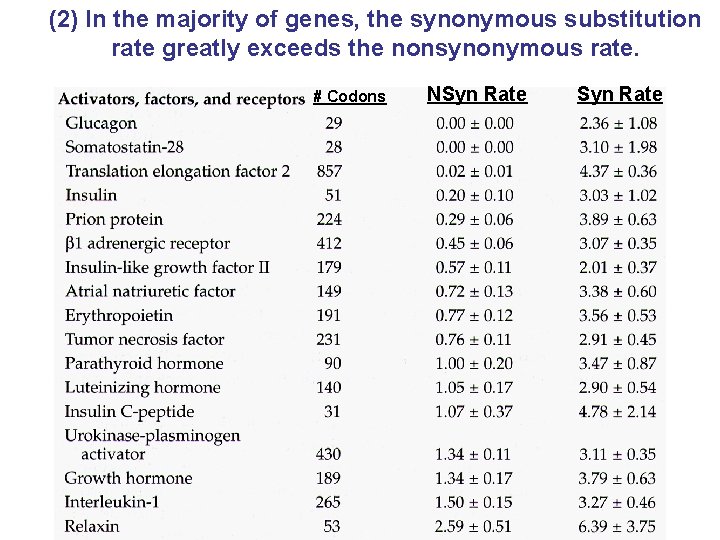
(2) In the majority of genes, the synonymous substitution rate greatly exceeds the nonsynonymous rate. # Codons NSyn Rate
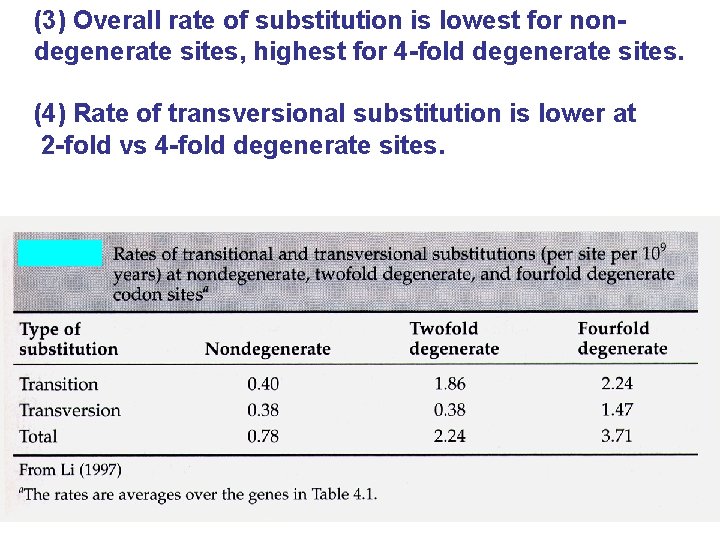
(3) Overall rate of substitution is lowest for nondegenerate sites, highest for 4 -fold degenerate sites. (4) Rate of transversional substitution is lower at 2 -fold vs 4 -fold degenerate sites.
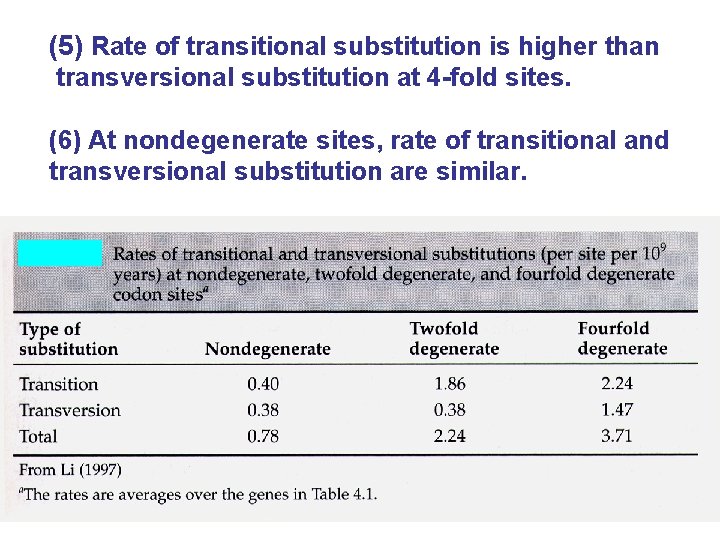
(5) Rate of transitional substitution is higher than transversional substitution at 4 -fold sites. (6) At nondegenerate sites, rate of transitional and transversional substitution are similar.
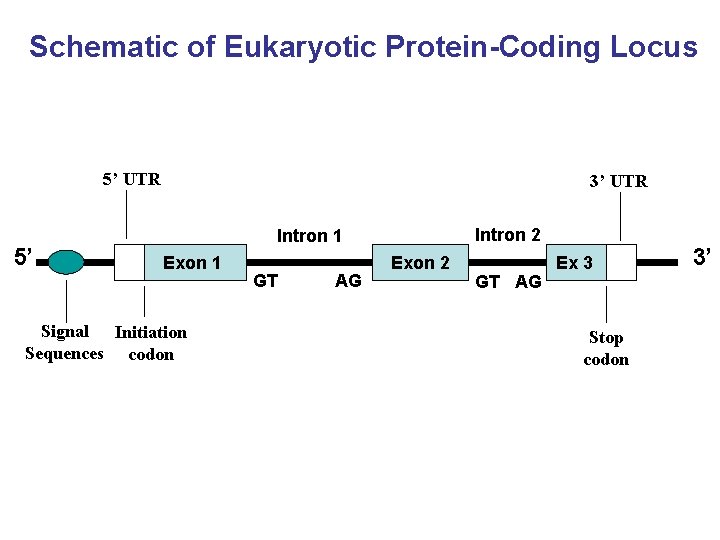
Schematic of Eukaryotic Protein-Coding Locus 5’ UTR 5’ 3’ UTR Intron 2 Intron 1 Exon 1 Signal Initiation Sequences codon GT AG Exon 2 GT AG Ex 3 Stop codon 3’
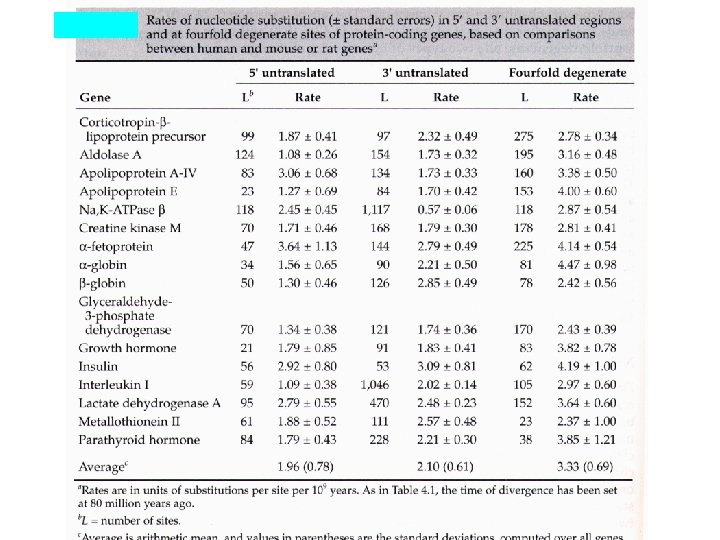
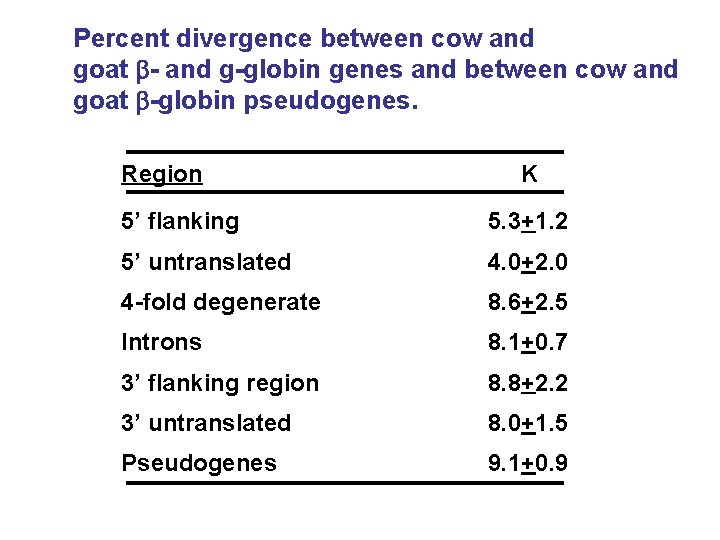
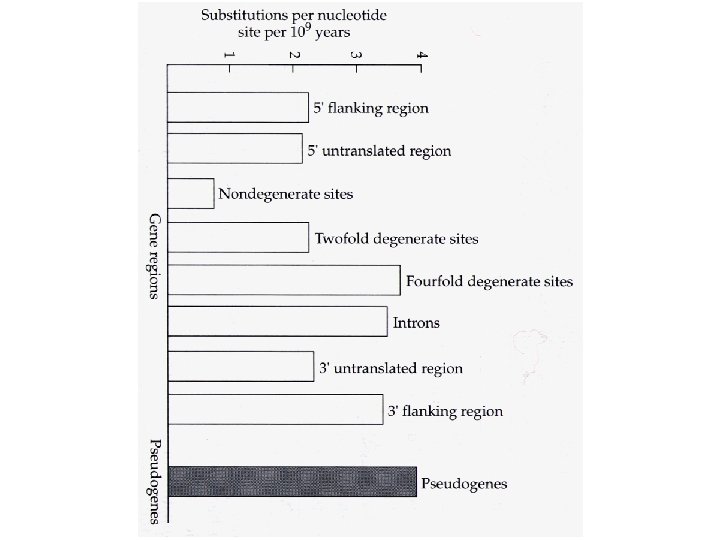
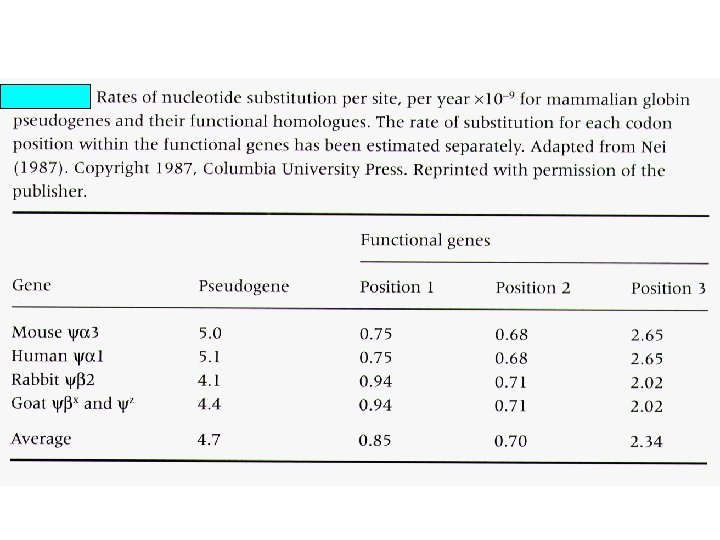
Percent divergence between cow and goat b- and g-globin genes and between cow and goat b-globin pseudogenes. Region K 5’ flanking 5. 3+1. 2 5’ untranslated 4. 0+2. 0 4 -fold degenerate 8. 6+2. 5 Introns 8. 1+0. 7 3’ flanking region 8. 8+2. 2 3’ untranslated 8. 0+1. 5 Pseudogenes 9. 1+0. 9
Causes of Variation in Substitution Rates Rate of Substitution is determined by: (1) Mutation rate Among genes Among gene regions (2) Probability of fixation Neutral, advantageous, deleterious
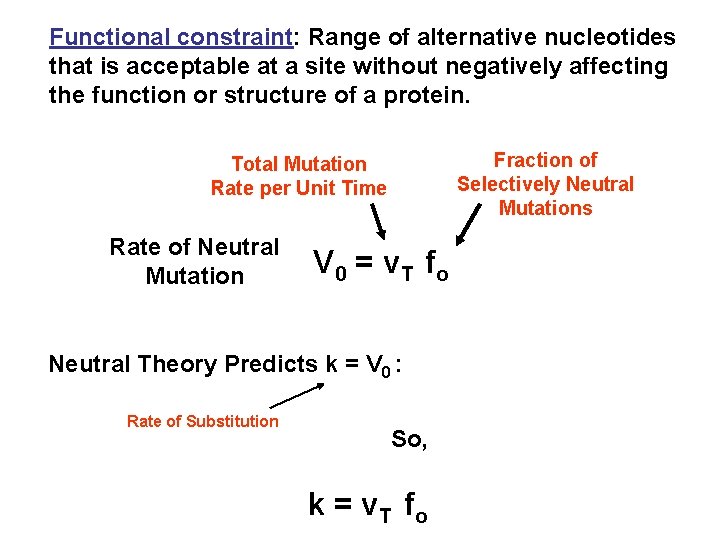
Functional constraint: Range of alternative nucleotides that is acceptable at a site without negatively affecting the function or structure of a protein. Fraction of Selectively Neutral Mutations Total Mutation Rate per Unit Time Rate of Neutral Mutation V 0 = v T fo Neutral Theory Predicts k = V 0 : Rate of Substitution So, k = v T fo
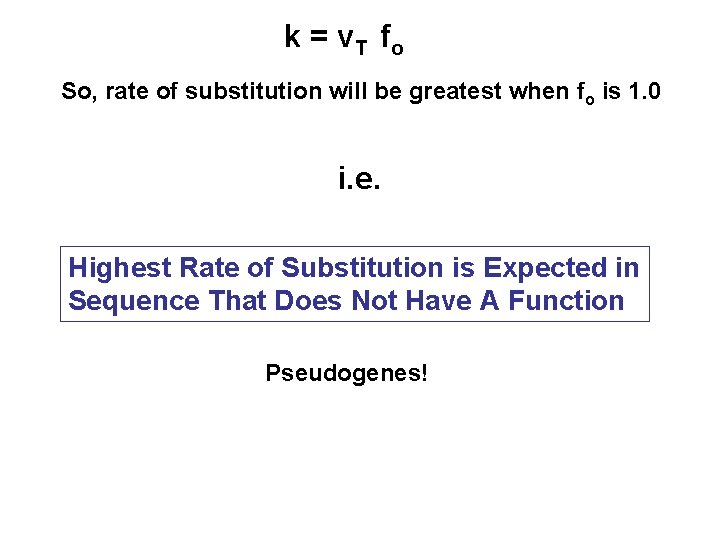
k = v T fo So, rate of substitution will be greatest when fo is 1. 0 i. e. Highest Rate of Substitution is Expected in Sequence That Does Not Have A Function Pseudogenes!
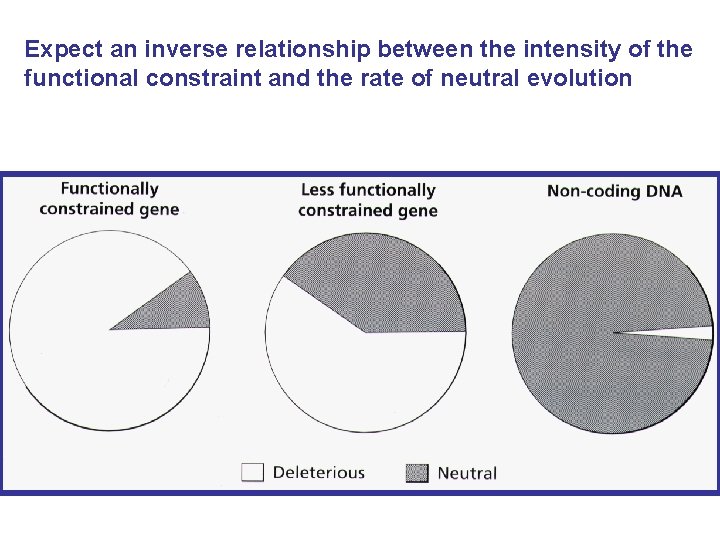
Expect an inverse relationship between the intensity of the functional constraint and the rate of neutral evolution

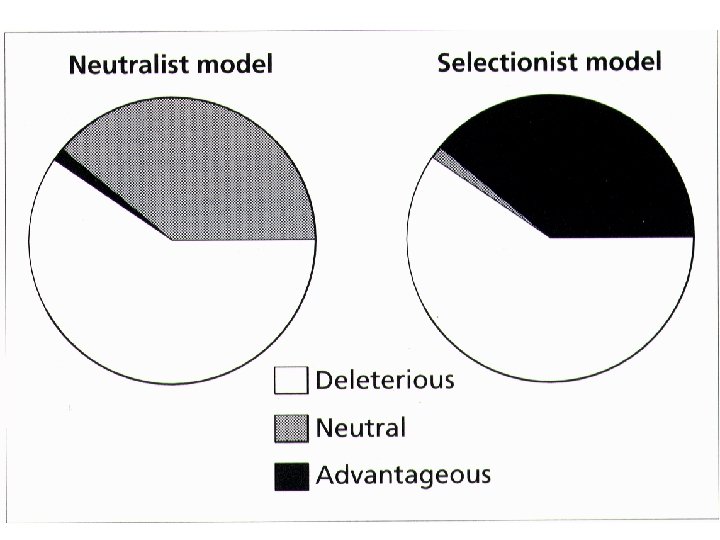
Given this relationship: Also, expect higher rates of substitution for synonymous vs nonsynonymous sites. Logic: (1) Mutations that result in amino acid replacements have a higher probability of causing a deleterious effect on the structure/function of the protein. (2) Accordingly, the majority of nonsynonomous mutations will be eliminated from the population by purifying selection. (3) As a result, there will be a reduction in the rate of nonsynonymous substitution vs synonymous substitution.

Causes of Variation in Substitution Rates Rate of Substitution is determined by: (1) Mutation rate Among genes Among gene regions (2) Probability of fixation Neutral, advantageous, deleterious
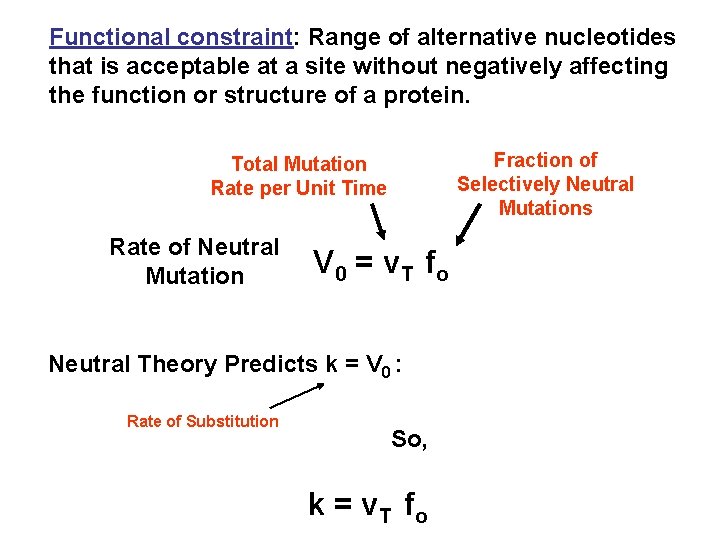
Functional constraint: Range of alternative nucleotides that is acceptable at a site without negatively affecting the function or structure of a protein. Fraction of Selectively Neutral Mutations Total Mutation Rate per Unit Time Rate of Neutral Mutation V 0 = v T fo Neutral Theory Predicts k = V 0 : Rate of Substitution So, k = v T fo
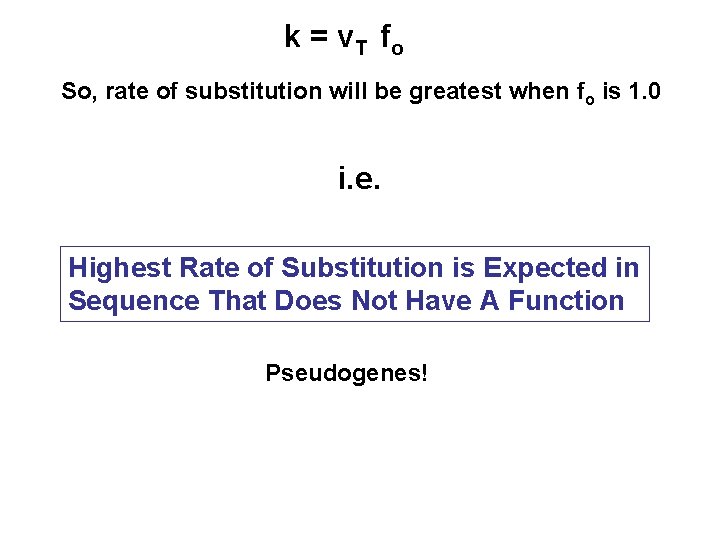
k = v T fo So, rate of substitution will be greatest when fo is 1. 0 i. e. Highest Rate of Substitution is Expected in Sequence That Does Not Have A Function Pseudogenes!
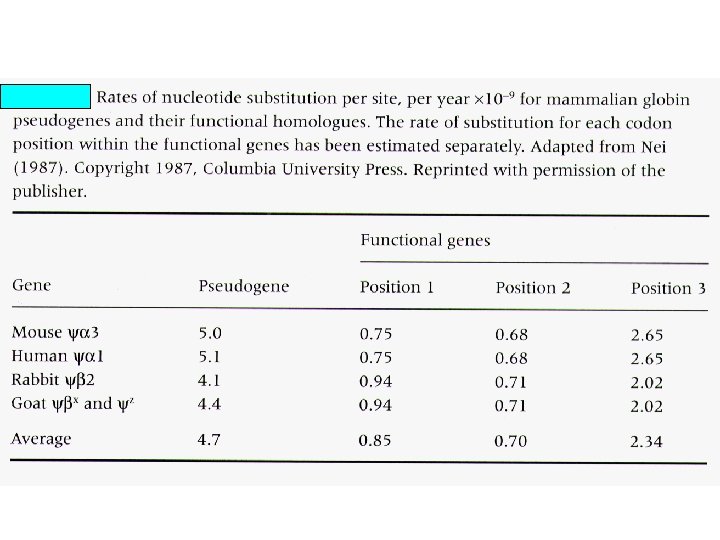
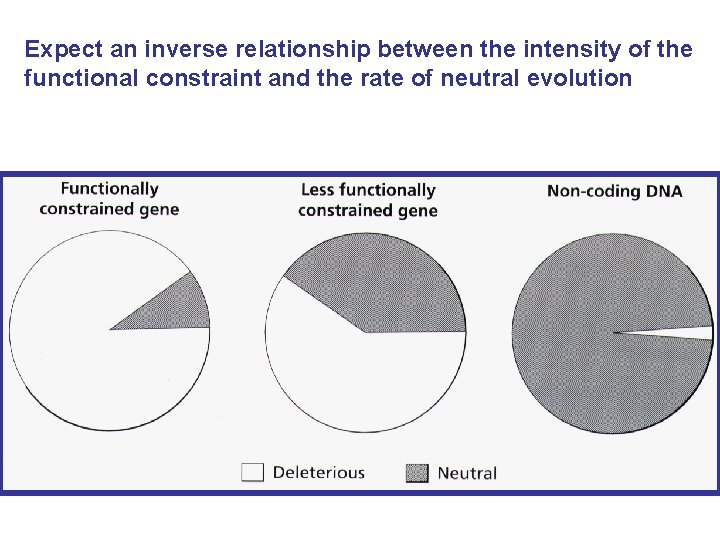
Expect an inverse relationship between the intensity of the functional constraint and the rate of neutral evolution

Given this relationship: Also, expect higher rates of substitution for synonymous vs nonsynonymous sites. Logic: (1) Mutations that result in amino acid replacements have a higher probability of causing a deleterious effect on the structure/function of the protein. (2) Accordingly, the majority of nonsynonomous mutations will be eliminated from the population by purifying selection. (3) As a result, there will be a reduction in the rate of nonsynonymous substitution vs synonymous substitution.
Why is the rate of substitution at 4 -fold sites lower than the rate within pseudogenes? Synonymous substitutions are not selectively neutral! Codon Usage is non-random: species-specific, and patterns may vary among genes within a genome.
- Slides: 24