Welcome to class of DNA Dr Meera Kaur















- Slides: 47

Welcome to class of DNA Dr. Meera Kaur

Nucleic Acid Structure • Structure of DNA: – The most important clue to the structure of DNA came from the work of Erwin Chargaff and his colleagues in the late 1940 s. – The data collected from DNAs of a great many different species led Chargaff to some important conclusions. These conclusions are as follows…

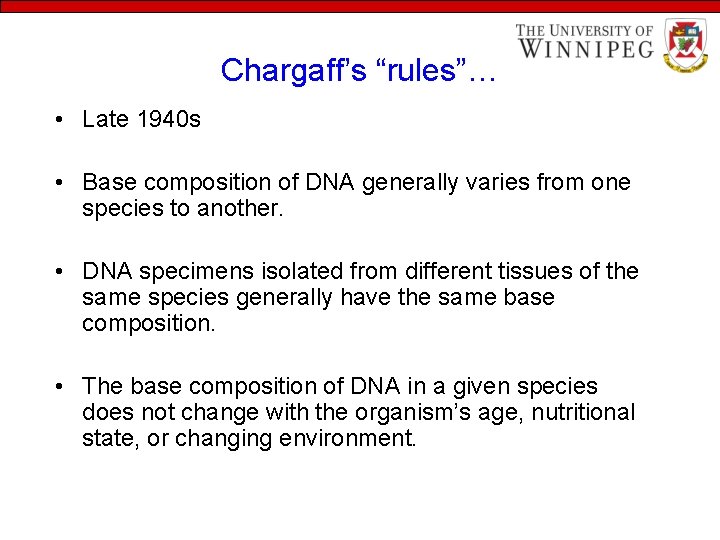
Chargaff’s “rules”… • Late 1940 s • Base composition of DNA generally varies from one species to another. • DNA specimens isolated from different tissues of the same species generally have the same base composition. • The base composition of DNA in a given species does not change with the organism’s age, nutritional state, or changing environment.

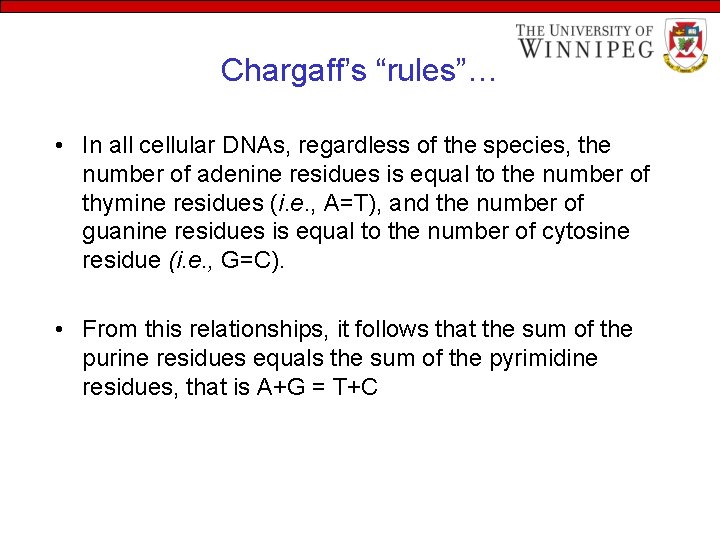
Chargaff’s “rules”… • In all cellular DNAs, regardless of the species, the number of adenine residues is equal to the number of thymine residues (i. e. , A=T), and the number of guanine residues is equal to the number of cytosine residue (i. e. , G=C). • From this relationships, it follows that the sum of the purine residues equals the sum of the pyrimidine residues, that is A+G = T+C

Chargaff’s “rules” • Key to establishing the 3 D structure of DNA • Provide clues as to how genetic information is encoded in DNA and passed from one generation to the next.

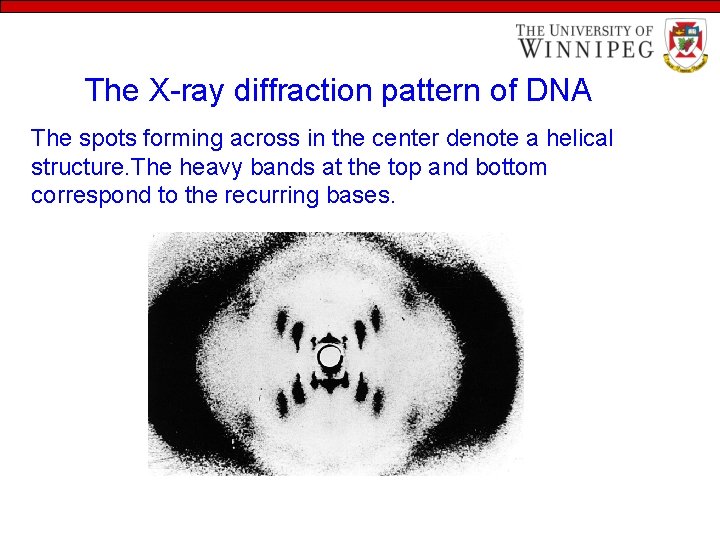
DNA is a double helx • 1950 s: Rosalind Franklin and Maurice Wilkins obtained X-ray diffraction pattern of DNA – deduced that DNA was helical. • The pattern also indicated that the molecule contained two strands, a clue that was crucial to determining the structure.

The X-ray diffraction pattern of DNA The spots forming across in the center denote a helical structure. The heavy bands at the top and bottom correspond to the recurring bases.

Race was on to determine structure • The problem was to formulate a threedimensional model of DNA molecule that could account not only the X-ray diffraction data, but also – Needed to be a helical structure, also satisfy Chargoff’s rules (A=T, C=G) and - Satisfied other chemical properties of DNA

Watson and Crick model for the structure of DNA • In 1953, Watson and Crick postulated that, – It consists of two helical DNA chains coiled around the same axis to form a right-handed double helix. The strands of DNA comprising the double helix run in opposite direction – The spatial relationship between the two strands creates a major groove and a minor groove between the two strands – The three - dimensional model of DNA structure confirms the Chargaff’s rules

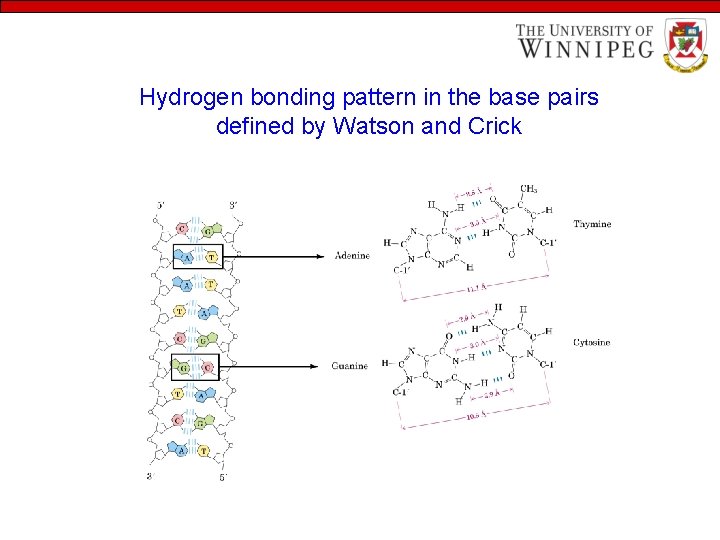
Hydrogen bonding pattern in the base pairs defined by Watson and Crick

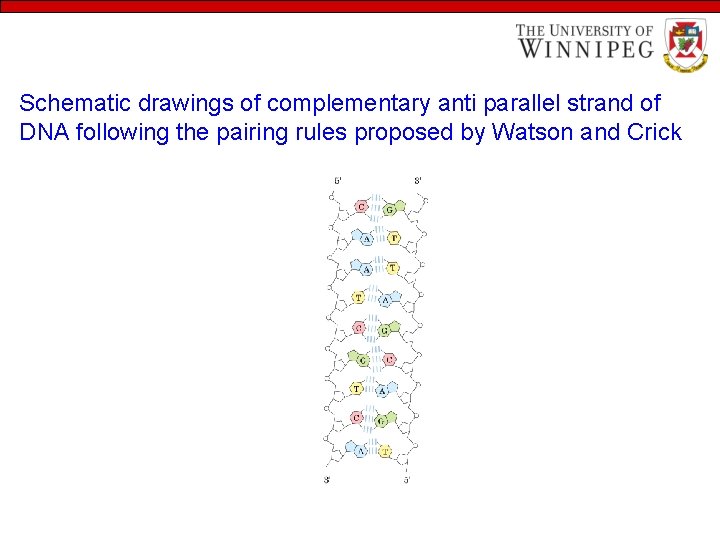
Schematic drawings of complementary anti parallel strand of DNA following the pairing rules proposed by Watson and Crick

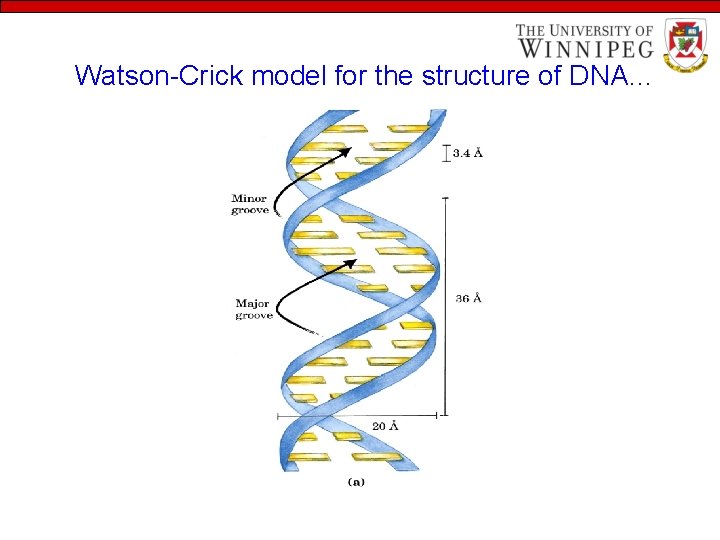
Watson-Crick model for the structure of DNA…

Watson-Crick model for the structure of DNA… • Watson and Crick postulated that DNA is a double helix where - Two helical DNA strands coiled around same axis to form right-handed double helix - Hydrophilic backbone of alternating sugar/phosphate units of the nucleotides are on outside of DNA molecule exposed to water

Watson-Crick model for the structure of DNA… • Hydrophobic bases (complementary base pairs) are stacked neatly inside the molecule. The hydrogen bond between the base pairs hold the double helix together. • Strands are anti parallel.

Watson-Crick model for the structure of DNA Note That • The two DNA strands in the “double helix” are anti parallel. • The two DNA strands are not IDENTICAL, but they are COMPLEMENTARY.

Important features of the double helical model of DNA • The model explained many unusual physical properties of DNA. • It immediately suggested a mechanism for the transmission of genetic information. • It suggested a means by which DNA could be replicated.

The central dogma outlines the plan for the storage and transmission of hereditary information. Now we know that DNA is the bearer of genetic information in all living organisms. The entire basis of information storage and transmission in the cell is embodied in three steps: REPLICATION TRANSCRIPTION TRANSLATION

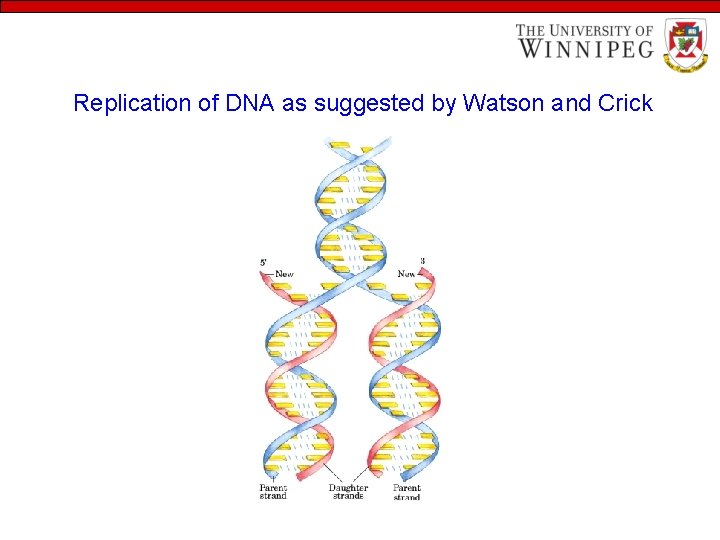
Replication Watson and Crick model for DNA replication They suggested that DNA is replicated on a DNA template. - Each strand of DNA in the double helix acts as a template – a pattern for the synthesis of its complement. Since DNA is double-stranded, complementary replication would produce two double-helical DNA molecules, each containing a strand of the original DNA and a new strand complementary to it. - This type of replication is called semiconservative replication, which was confirmed by M. S. Meselson and F. W. Stahl in 1957. - In 1955, Arthur Kornberg and his associates discovered DNA polymerase — a large protein which catalyses the formation of DNA. - DNA strand elongates. Elongation or the chain growth is always from the 5 end to the 3 end of the elongating DNA molecules

Replication of DNA as suggested by Watson and Crick

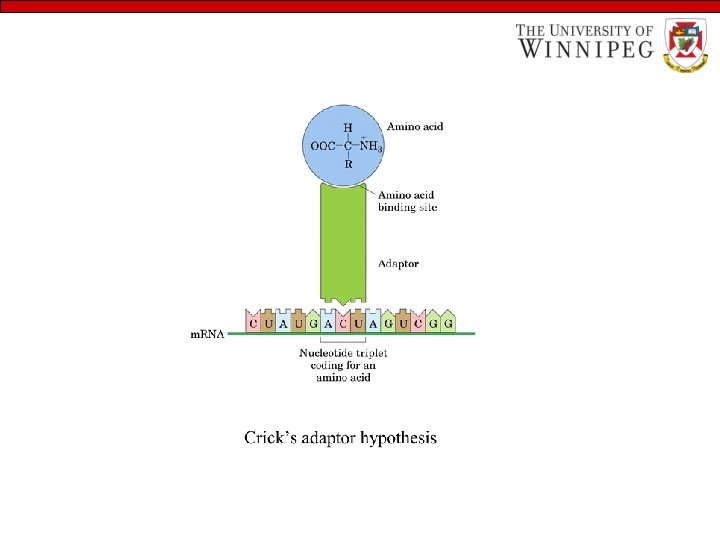
Some basics • The amino acids sequence of every protein and nucleotides sequence of every RNA molecule in cell is specified by that cell’s DNA. • A segment of DNA that contains the information required for the synthesis of a functional biological product is referred to as a GENE. • Ribosomal RNAs (r RNA) are the structural component of ribosome, the large complexes carry out the synthesis of protein. • Messenger RNAs (m. RNA) are nucleic acids that carry the information from one or a few genes to the ribosome, where the corresponding protein can be synthesized. • Transfer RNAs (t. RNA) are adaptor molecules that faithfully translate the information in m. RNA into a specific sequence of amino acids.

Transcription RNA is made on a DNA template • The information contained in DNA is passed to a form of RNA called messenger RNA (m. RNA) by transcription (“rewriting”). • The mechanics of transcription is quite similar to the mechanics of replication because DNA is the template upon which RNA is formed. The major difference is that— - The enzyme is RNA polymerase instead of DNA polymerase - Sugar units of the nucleoside used to make RNA is ribose rather than deoxyribose, and the u (uracil) substitutes for T (Thymidine) in RNA.

Translation involves t. RNA, m. RNA, ribosome and Enzymes The synthesis of specific protein under the direction of specific gene is complex. Proteins are the polymer of 20 different amino acids and there are only four different nucleotide monomers in DNA. Hence, there can not be a one-to-one relationship between the sequence of nucleotides in the DNA molecule and the sequence of amino acids in a protein. The protein-coding information is read by the cells in blocks of three nucleotides residues, or codons. Each codon specifies a different amino acids. The set of rules that specifies which nucleic acid codon corresponds to which amino acid is known as genetic code.


The basic principles of Translation – A m. RNA molecule is bound to a ribosome. – The transfer RNA(t. RNA) molecules bring amino acids to the ribosome one at a time. – Each t. RNA identifies the appropriate codon on the m. RNA and add this amino acid to the growing protein chain. – The first t. RNA is released and the ribosome moves one codon length along the message, allowing the next t. RNA to come into place, carrying its amino acid. Energy in the form of ATP is required at each step in the movement. – As the ribosome moves along the m. RNA , it eventually encounters a ‘stop’ codon. At this point, the polypeptide chain is released.

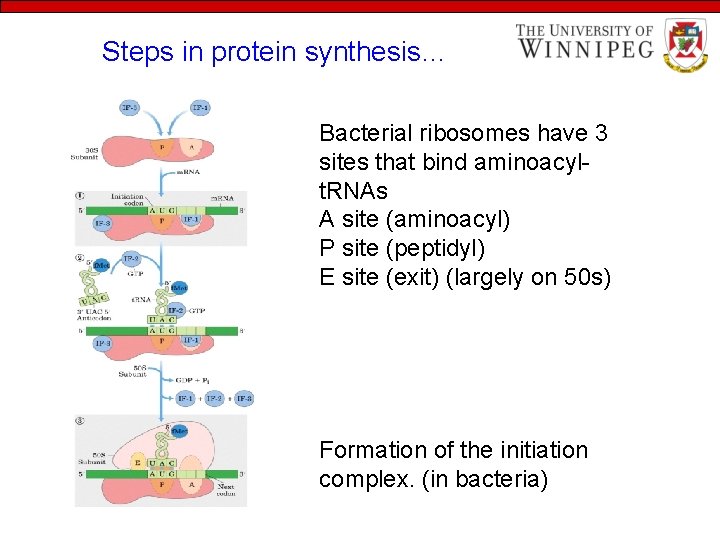
Steps in protein synthesis… Bacterial ribosomes have 3 sites that bind aminoacylt. RNAs A site (aminoacyl) P site (peptidyl) E site (exit) (largely on 50 s) Formation of the initiation complex. (in bacteria)

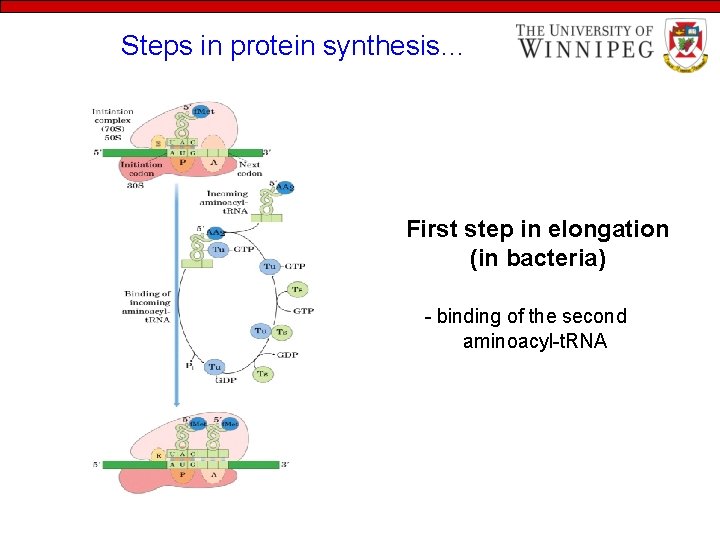
Steps in protein synthesis… First step in elongation (in bacteria) - binding of the second aminoacyl-t. RNA

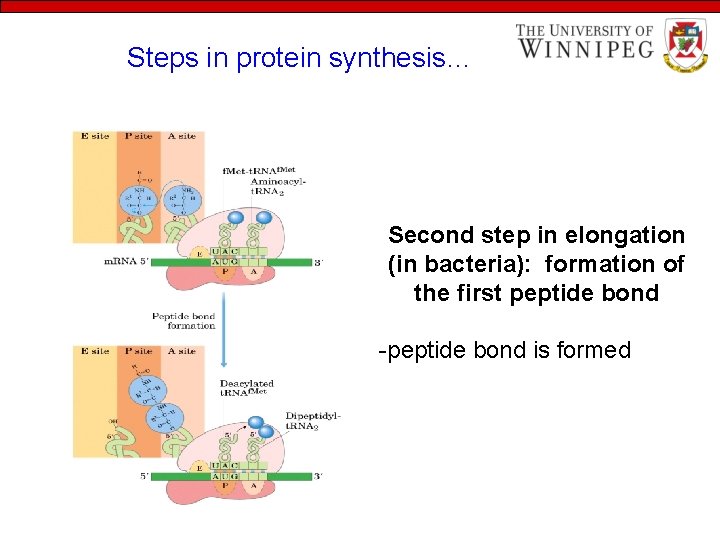
Steps in protein synthesis… Second step in elongation (in bacteria): formation of the first peptide bond -peptide bond is formed

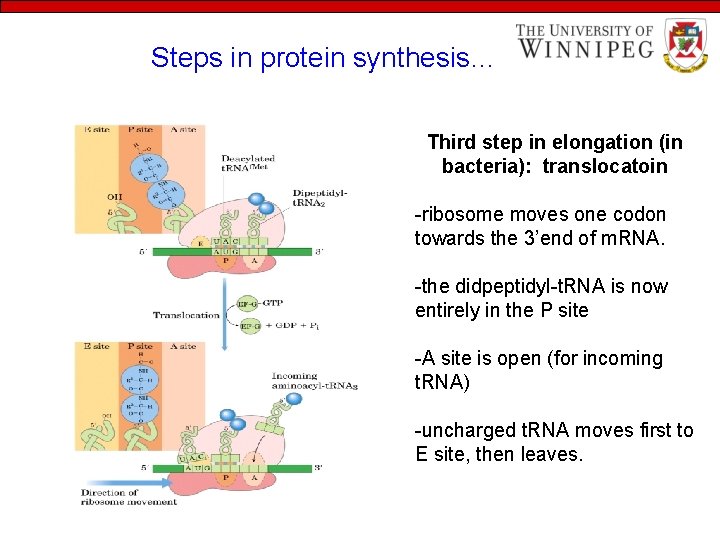
Steps in protein synthesis… Third step in elongation (in bacteria): translocatoin -ribosome moves one codon towards the 3’end of m. RNA. -the didpeptidyl-t. RNA is now entirely in the P site -A site is open (for incoming t. RNA) -uncharged t. RNA moves first to E site, then leaves.

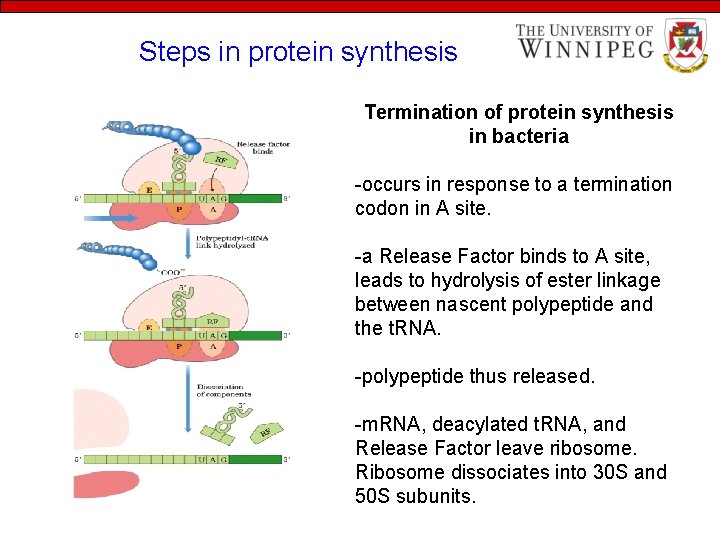
Steps in protein synthesis Termination of protein synthesis in bacteria -occurs in response to a termination codon in A site. -a Release Factor binds to A site, leads to hydrolysis of ester linkage between nascent polypeptide and the t. RNA. -polypeptide thus released. -m. RNA, deacylated t. RNA, and Release Factor leave ribosome. Ribosome dissociates into 30 S and 50 S subunits.

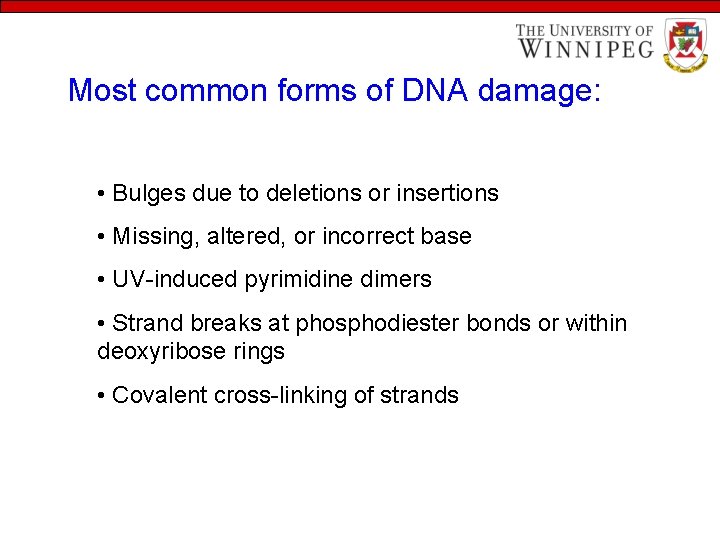
Most common forms of DNA damage: • Bulges due to deletions or insertions • Missing, altered, or incorrect base • UV-induced pyrimidine dimers • Strand breaks at phosphodiester bonds or within deoxyribose rings • Covalent cross-linking of strands

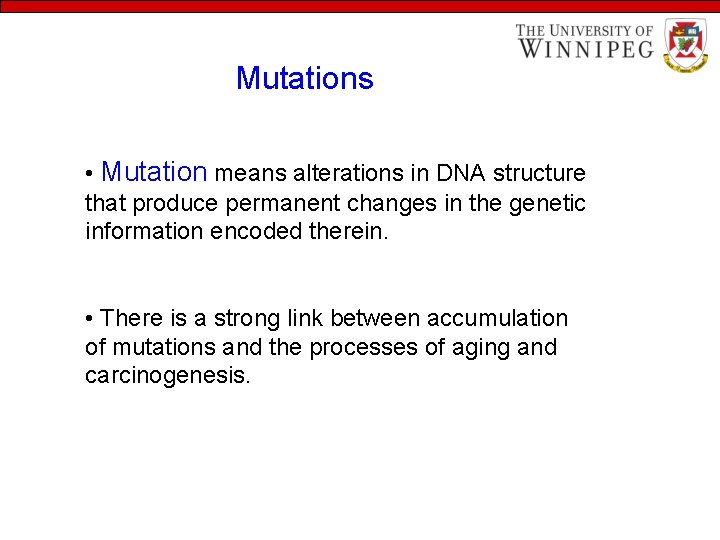
Mutations • Mutation means alterations in DNA structure that produce permanent changes in the genetic information encoded therein. • There is a strong link between accumulation of mutations and the processes of aging and carcinogenesis.

Recombinant DNA • There are no cure for molecular diseases. But medical scientists dream to correct these inborn errors by replacing a nonfunctional gene on a human chromosome with one that is functional. The research involves experiments with recombinant DNA. • Recombination consists of cleaving DNA chains, inserting a new piece of DNA into the gap created by the cleavage, and resealing the chains.

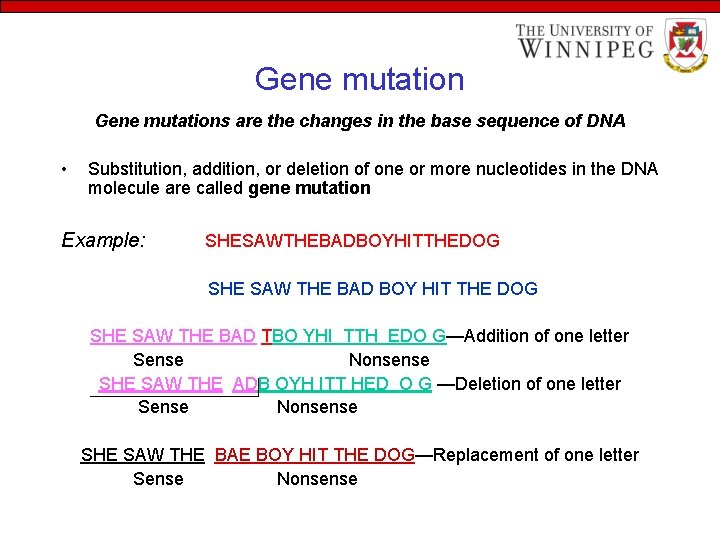
Gene mutations are the changes in the base sequence of DNA • Substitution, addition, or deletion of one or more nucleotides in the DNA molecule are called gene mutation Example: SHESAWTHEBADBOYHITTHEDOG SHE SAW THE BAD BOY HIT THE DOG SHE SAW THE BAD TBO YHI TTH EDO G—Addition of one letter Sense Nonsense SHE SAW THE ADB OYH ITT HED O G —Deletion of one letter Sense Nonsense SHE SAW THE BAE BOY HIT THE DOG—Replacement of one letter Sense Nonsense

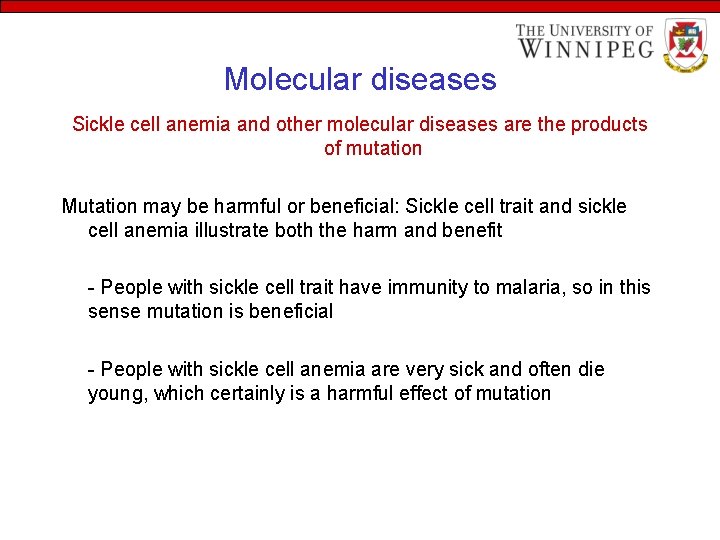
Molecular diseases Sickle cell anemia and other molecular diseases are the products of mutation Mutation may be harmful or beneficial: Sickle cell trait and sickle cell anemia illustrate both the harm and benefit - People with sickle cell trait have immunity to malaria, so in this sense mutation is beneficial - People with sickle cell anemia are very sick and often die young, which certainly is a harmful effect of mutation

Xeroderma pigmentosum (XP) • Disease caused by defect in the nucleotide-excision repair system. • People with XP are extremely light sensitive, and readily develop sunlight-induced cancers. • They also have neurological abnormalities, presumably due to inability to repair certain lesions caused by high rate of oxidative metabolism in neurons

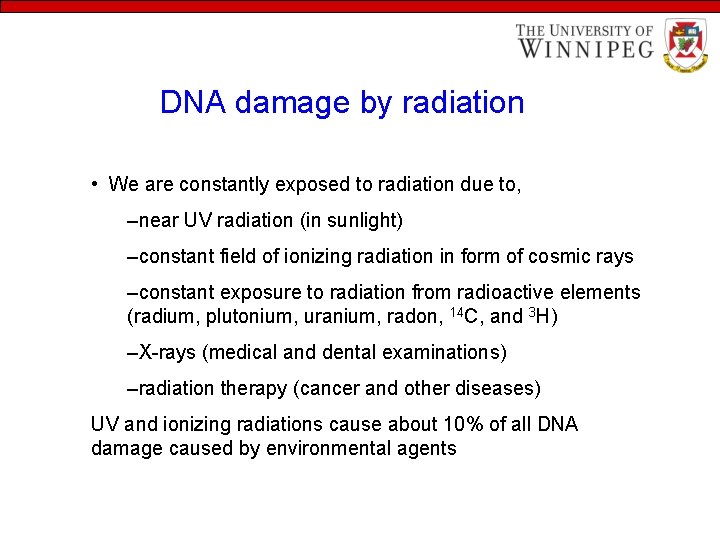
DNA damage by radiation • We are constantly exposed to radiation due to, –near UV radiation (in sunlight) –constant field of ionizing radiation in form of cosmic rays –constant exposure to radiation from radioactive elements (radium, plutonium, uranium, radon, 14 C, and 3 H) –X-rays (medical and dental examinations) –radiation therapy (cancer and other diseases) UV and ionizing radiations cause about 10% of all DNA damage caused by environmental agents

Oxidative damage to DNA… • Oxidative damage is possibly the most important source of mutagenic alterations in DNA. • Excited oxygen species such as hydrogen peroxide, hydroxyl radicals, and superoxide radicals arise during irradiation or as a byproduct of aerobic metabolism

Oxidative damage to DNA • Cells have elaborate defense system to destroy these reactive species. Thses includes enzymes catalase and superoxide dismutase that convert reactive oxygen species to harmless products. • A fraction of these species will escape cellular defenses, and cause damage (ranging from oxidation of deoxyribose and base moieties to strand breaks) • Data related to this type of damage is not available. However, every day DNA of each human cell exposed to thousands of such reactions.

Hereditary nonpolyposis colon cancer (HNPCC) • This type of cancer generally develops at an early age. • This is caused by defects in mismatch repair. Defects in at least five different mismatch repair genes can give rise to HNPCC

Breast cancer • Mostly occurs in women with no known predisposition. • At least 10% of cases are associated with defects in two genes (Brca 1 and Brca 2) associated with DNA repair. • Women with defects in either of these genes have a >80% chance of developing breast cancer.

DNA repair… • Biological macromolecules are susceptible to chemical alterations that arise from environmental damage or errors during synthesis. • For RNAs, proteins, or other cellular molecules, most consequences of such damage are circumvented through normal turnover (synthesis and degradation).

DNA repair… • However, integrity of DNA is vital to cell survival and reproduction. • Its information content must be protected over the life span of the cell and preserved from generation to generation.

DNA repair… • DNA is the only molecule that, if damaged, is repaired by the cell. • Such repair is possible because the information content of duplex DNA is inherently redundant.

DNA repair… • Human DNA replication has an error rate of about three base-pair mistakes during copying of 6 billion base pairs in the diploid human genome. - low error rate is due to DNA repair systems that review and edit the newly replicated DNA • Firther, about 10, 000 bases (mostly purines) are lost per cell per day from spontaneous breakdown in human DNA; the repair systems must replace these bases to maintain the fidelity of the encoded information.

DNA repair… • Usually, the complementary structure of DNA ensures that the information lost through damage to one strand can be recovered from the other. • However, even errors involving both strands can be corrected through recombination. • Double-stranded breaks (potentially the most serious lesions) can be repaired by recombination events.

Nucleic Acid Chemistry • Double – helical DNA and RNA can be denatured (extreme p. H and heat) • Nucleic acids from different species can form hybrids • Nucleotides and nucleic acids undergo non enzymatic browning • DNA is often methylated • Long DNA sequences can be determined

Other functions of nucleotides • Nucleotides carry chemical energy in cells. • Nucleotides are components of many enzyme cofactors. • Some nucleotides are intermediates in cellular communications.
 Meera kaur
Meera kaur Meera kaur
Meera kaur Watson crick model of dna
Watson crick model of dna Meera kaur
Meera kaur Manisha has secured 79 marks in the college exam
Manisha has secured 79 marks in the college exam Replication
Replication Bioflix activity dna replication nucleotide pairing
Bioflix activity dna replication nucleotide pairing Coding dna and non coding dna
Coding dna and non coding dna What are the enzymes involved in dna replication
What are the enzymes involved in dna replication Chapter 11 dna and genes
Chapter 11 dna and genes Meera mahajan
Meera mahajan Meera mahajan
Meera mahajan Meera warrier
Meera warrier Dr meera sood
Dr meera sood Meera warrier
Meera warrier Meera venkatesh
Meera venkatesh Jaiya gill
Jaiya gill Acknowledgement for project
Acknowledgement for project Mata bhag kaur
Mata bhag kaur Gulnaz kaur
Gulnaz kaur Bus topology advantages and disadvantages
Bus topology advantages and disadvantages Adhesiivinen kuluminen
Adhesiivinen kuluminen Jasleen kaur unc
Jasleen kaur unc Rajkumari amrit kaur coaching scheme
Rajkumari amrit kaur coaching scheme Kaur kukkur
Kaur kukkur Vaido vahter
Vaido vahter Kaur kajak
Kaur kajak Pam kaur hsbc signature
Pam kaur hsbc signature Aare saar
Aare saar Rani sahib kaur
Rani sahib kaur Kaur aare saar
Kaur aare saar Cremation in sikhism
Cremation in sikhism Kaur aare saar
Kaur aare saar Aare saar
Aare saar Kaur aare saar
Kaur aare saar Pauliina heinänen
Pauliina heinänen Kaur jaakma
Kaur jaakma Kaur aare saar
Kaur aare saar Martin kaur
Martin kaur Manroop kaur
Manroop kaur Pruunkaru rahvapärased nimetused
Pruunkaru rahvapärased nimetused Tommi haapaniemi
Tommi haapaniemi Rajkumari amrit kaur coaching scheme
Rajkumari amrit kaur coaching scheme Harwinder dhillon
Harwinder dhillon Dr prabhdeep kaur
Dr prabhdeep kaur Kaur aare saar
Kaur aare saar Meaning of maninder kaur
Meaning of maninder kaur Welcome welcome this is our christmas story
Welcome welcome this is our christmas story