Welcome to BISC 220 Cell Physiology Lab Instructor

Welcome to BISC 220 Cell Physiology Lab Instructor: Jennifer Hood-De. Grenier Office: SC 376 A, x 3313 Research Lab: SC 311, x 3387 Email: jhooddeg@wellesley. edu Office Hours: Tues. 1: 30 -2: 30 pm Thurs. 9: 30 -10: 30 am Or e-mail to schedule an appointment
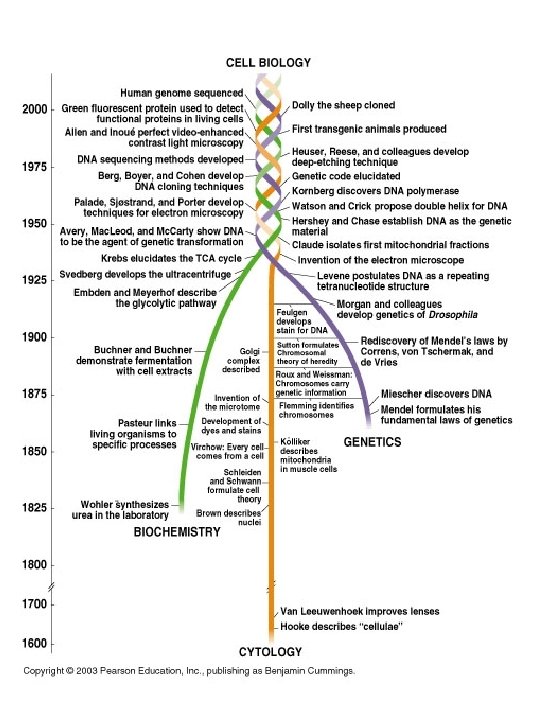
The Four Strands of Modern Cell Biology • Cytology: observation of cells by microscopy • Biochemistry: reductionist approach; in vitro study of biological molecules • Genetics: study of the effect of heritable information (DNA) on cell behavior/attributes; use of mutants to study cellular processes • Bioinformatics: application of computer algorithms to the analysis of large databases of biological information (genomics/proteomics)
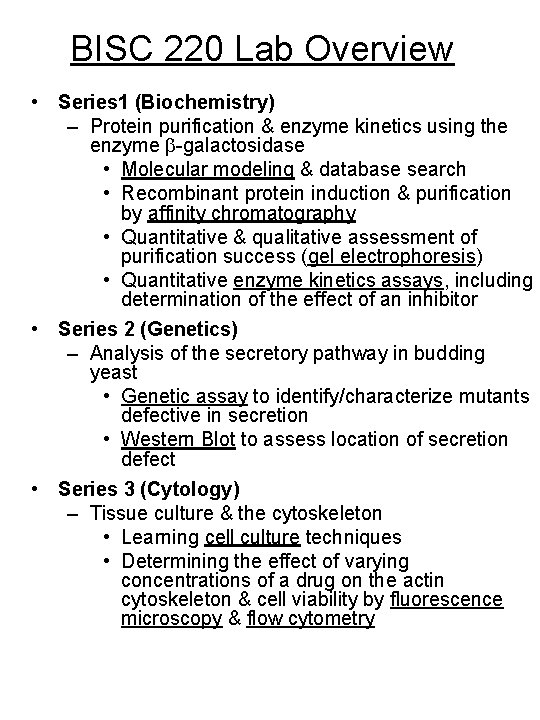
BISC 220 Lab Overview • Series 1 (Biochemistry) – Protein purification & enzyme kinetics using the enzyme -galactosidase • Molecular modeling & database search • Recombinant protein induction & purification by affinity chromatography • Quantitative & qualitative assessment of purification success (gel electrophoresis) • Quantitative enzyme kinetics assays, including determination of the effect of an inhibitor • Series 2 (Genetics) – Analysis of the secretory pathway in budding yeast • Genetic assay to identify/characterize mutants defective in secretion • Western Blot to assess location of secretion defect • Series 3 (Cytology) – Tissue culture & the cytoskeleton • Learning cell culture techniques • Determining the effect of varying concentrations of a drug on the actin cytoskeleton & cell viability by fluorescence microscopy & flow cytometry
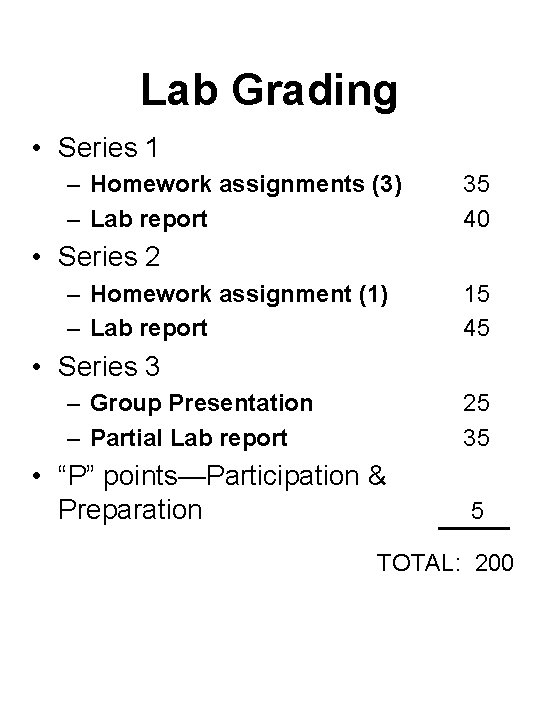
Lab Grading • Series 1 – Homework assignments (3) – Lab report 35 40 • Series 2 – Homework assignment (1) – Lab report 15 45 • Series 3 – Group Presentation – Partial Lab report 25 35 • “P” points—Participation & Preparation 5 TOTAL: 200
Lab 1 • Induction of -galactosidase ( gal) expression in E. coli • Ras. Mol – Investigation of the structure of -gal • Clustal. W – Identification of amino acid residues conserved among gal proteins from different species Next week: purification of -gal for study of its enzymatic properties
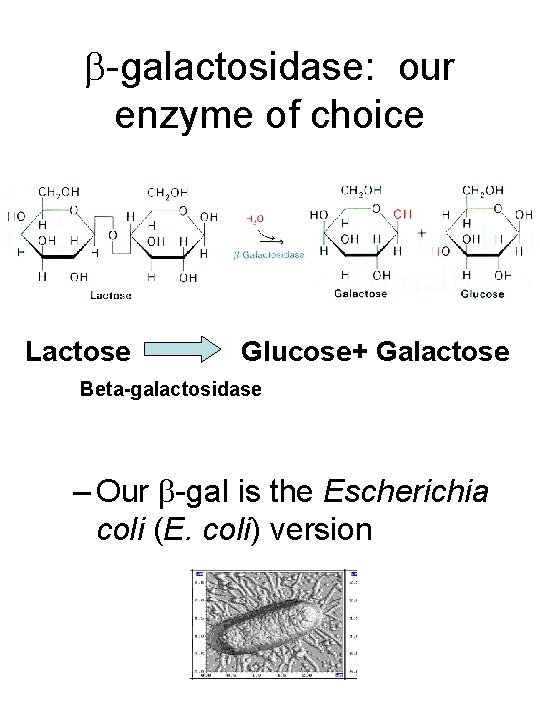
-galactosidase: our enzyme of choice Lactose Glucose+ Galactose Beta-galactosidase – Our -gal is the Escherichia coli (E. coli) version
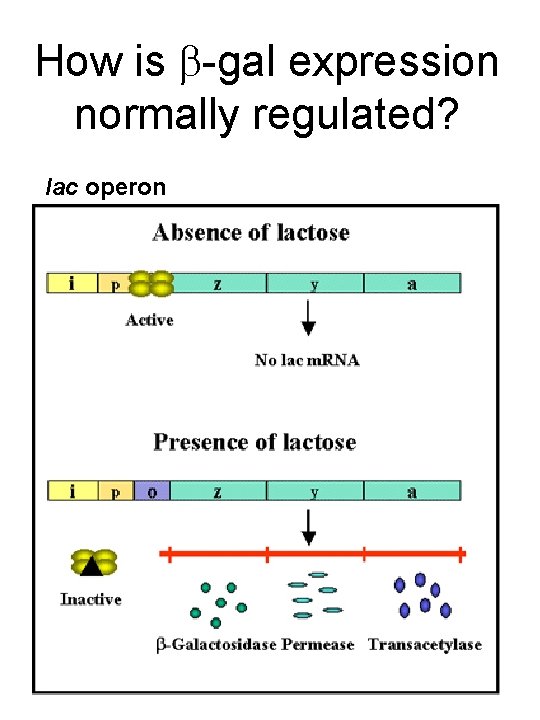
How is -gal expression normally regulated? lac operon
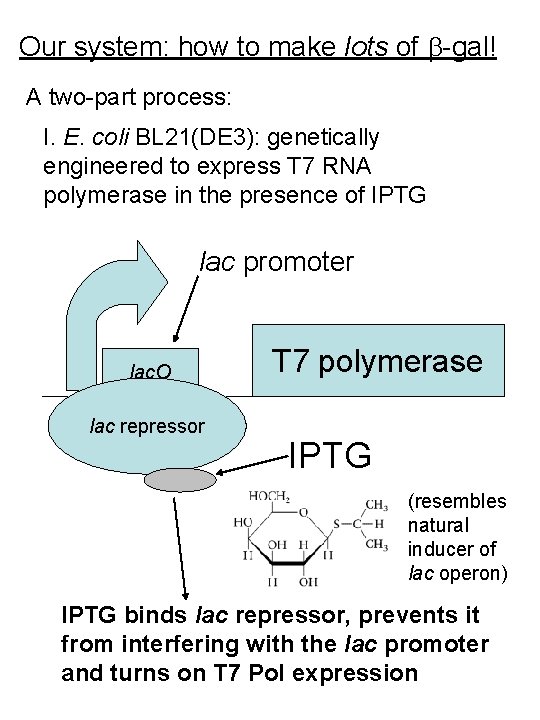
Our system: how to make lots of -gal! A two-part process: I. E. coli BL 21(DE 3): genetically engineered to express T 7 RNA polymerase in the presence of IPTG lac promoter lac. O lac repressor T 7 polymerase IPTG (resembles natural inducer of lac operon) IPTG binds lac repressor, prevents it from interfering with the lac promoter and turns on T 7 Pol expression
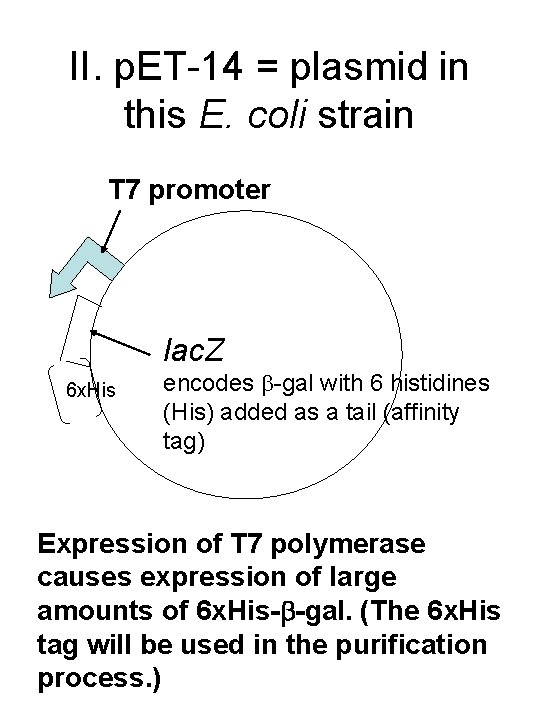
II. p. ET-14 = plasmid in this E. coli strain T 7 promoter lac. Z 6 x. His encodes -gal with 6 histidines (His) added as a tail (affinity tag) Expression of T 7 polymerase causes expression of large amounts of 6 x. His- -gal. (The 6 x. His tag will be used in the purification process. )

Protocol: Things to Remember • Think about aseptic technique (avoid contaminating your culture!) • Make flow chart of procedure and record all results in lab notebook • Do not discard anything contaminated with bacteria in sink—put growth medium in waste container or back in flask (must be treated with bleach) • Give labeled cell pellets to instructor freezing: – Pre-IPTG induction (small aliquot in tube) – After IPTG (remainder in centrifuge bottle)

While your bacteria are making lots of 6 x. His- -gal… • Calibrate micropipets • Look at CD animation of pdb file • Follow Ras. Mol tutorial in Appendix 1 Lab 1 (groups of 2) • Follow Clustal. W instructions in Appendix 2 Lab 1 (same groups as Ras. Mol)
A quick review of protein structure • Levels of structure: primary, secondary, tertiary & quaternary • Secondary structure elements: ahelices & -sheets • R-group interactions – Salt bridges (ionic interactions), Hydrogen bonds, van der Waals forces, hydrophobic interactions, disulfide bonds • Use of X-ray crystallography to “solve” protein structures (important for determining enzyme mechanisms, designing drugs, engineering mutations that alter protein function)
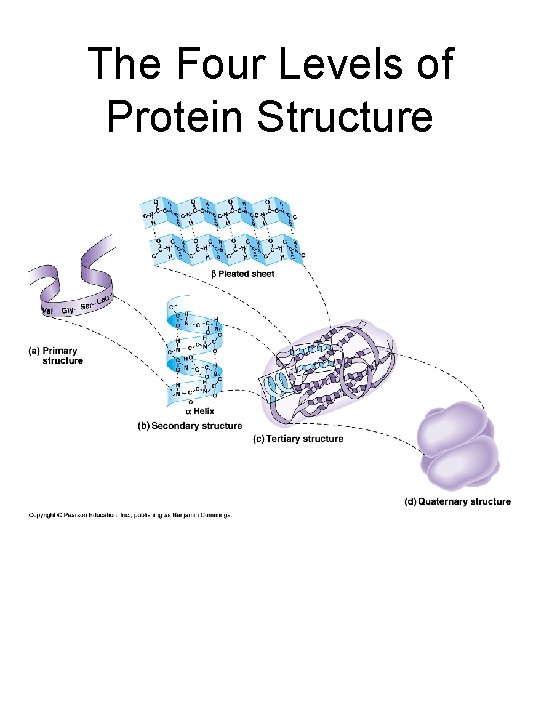
The Four Levels of Protein Structure
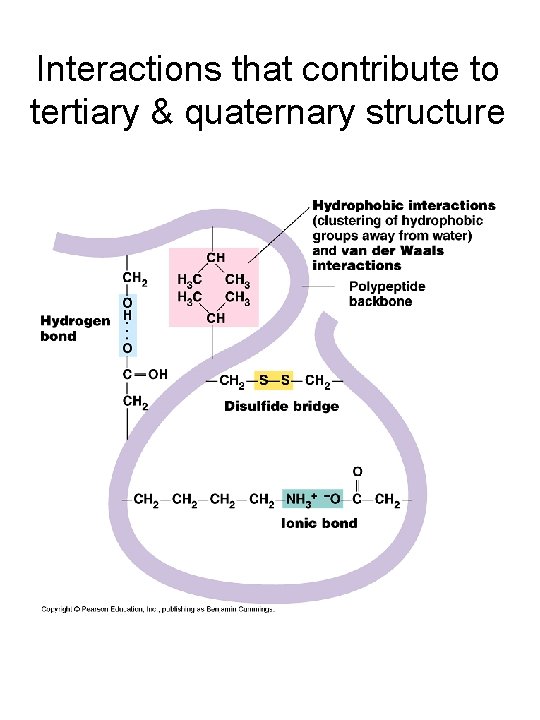
Interactions that contribute to tertiary & quaternary structure
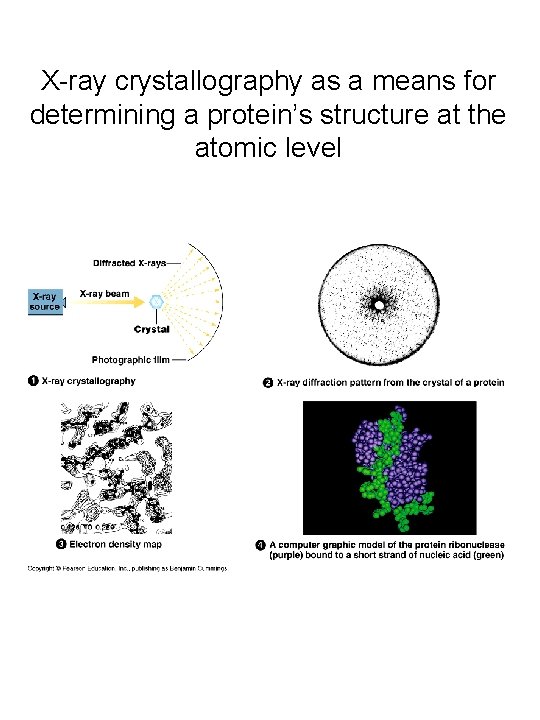
X-ray crystallography as a means for determining a protein’s structure at the atomic level

Homework • Do individually • Due next lab; 10 points • Create a figure with a correctly formatted legend from your saved Ras. Mol picture of the active site of -gal. • Include a paragraph (up to 1 page) describing what you learned about the active site. Try to relate the Clustal. W analysis to the structural analysis. • May consult references listed at end of HW assignment in lab manual.
- Slides: 17