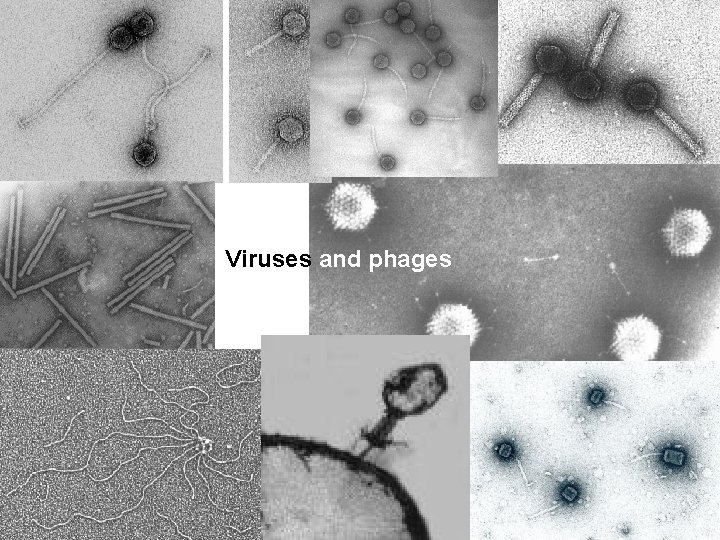
Viruses and phages Several types of independent genetic

- Slides: 53

Viruses and phages

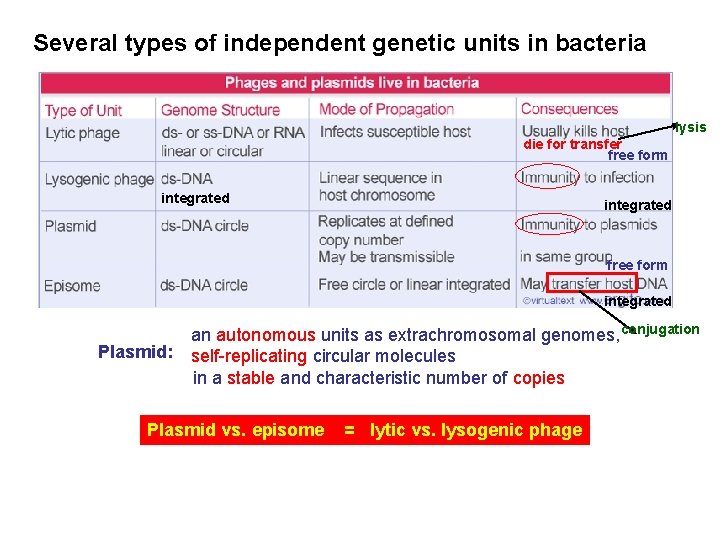
Several types of independent genetic units in bacteria lysis die for transfer free form integrated Plasmid: an autonomous units as extrachromosomal genomes, conjugation self-replicating circular molecules in a stable and characteristic number of copies Plasmid vs. episome = lytic vs. lysogenic phage

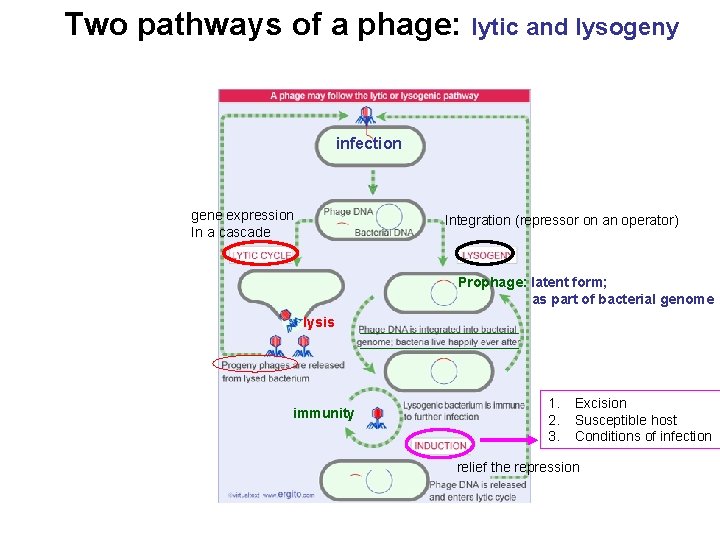
Two pathways of a phage: lytic and lysogeny infection gene expression In a cascade Integration (repressor on an operator) Prophage: latent form; as part of bacterial genome lysis immunity 1. 2. 3. Excision Susceptible host Conditions of infection relief the repression

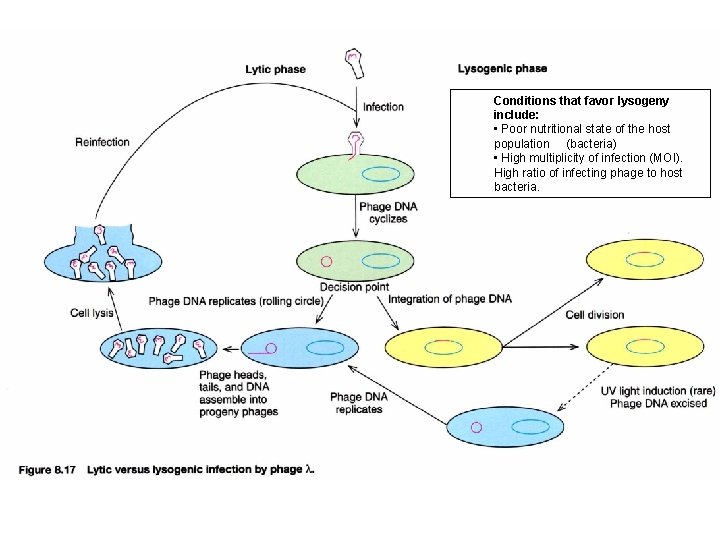
Conditions that favor lysogeny include: • Poor nutritional state of the host population (bacteria) • High multiplicity of infection (MOI). High ratio of infecting phage to host bacteria.

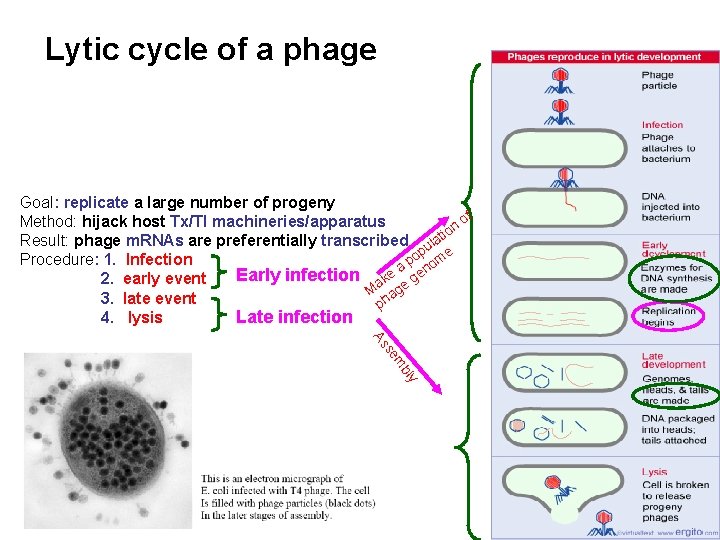
Lytic cycle of a phage Goal: replicate a large number of progeny of Method: hijack host Tx/Tl machineries/apparatus n o Result: phage m. RNAs are preferentially transcribed ulati p e Procedure: 1. Infection po om Early infection ake a gen 2. early event M age 3. late event ph 4. lysis Late infection bly em s As

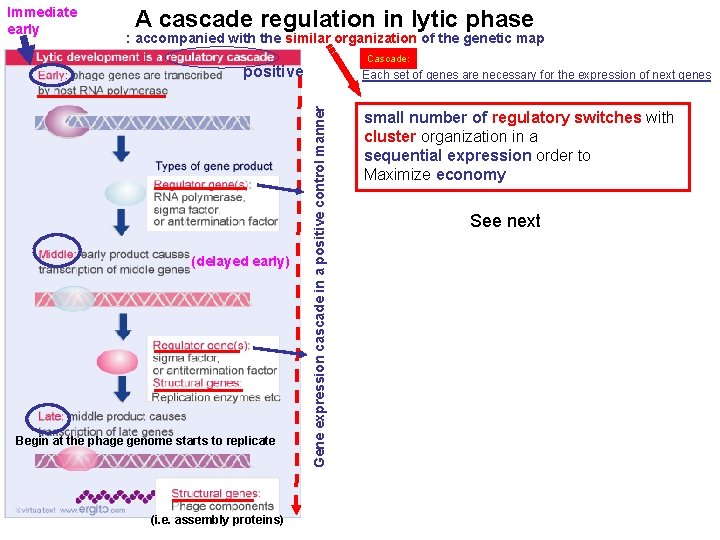
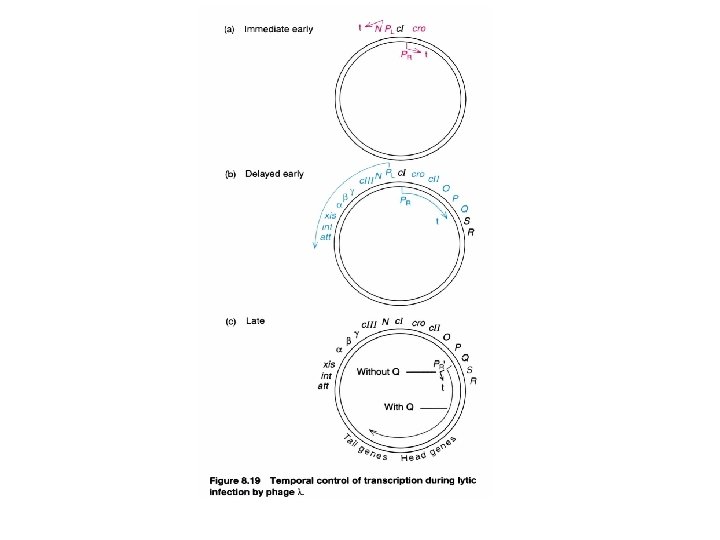
Immediate early A cascade regulation in lytic phase : accompanied with the similar organization of the genetic map Cascade: positive Begin at the phage genome starts to replicate (i. e. assembly proteins) Gene expression cascade in a positive control manner (delayed early) Each set of genes are necessary for the expression of next genes small number of regulatory switches with cluster organization in a sequential expression order to Maximize economy See next

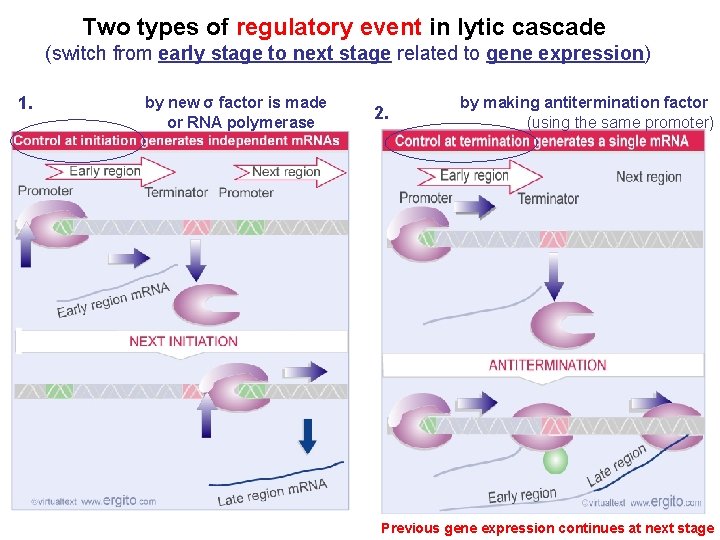
Two types of regulatory event in lytic cascade (switch from early stage to next stage related to gene expression) 1. by new σ factor is made or RNA polymerase 2. by making antitermination factor (using the same promoter) Previous gene expression continues at next stage

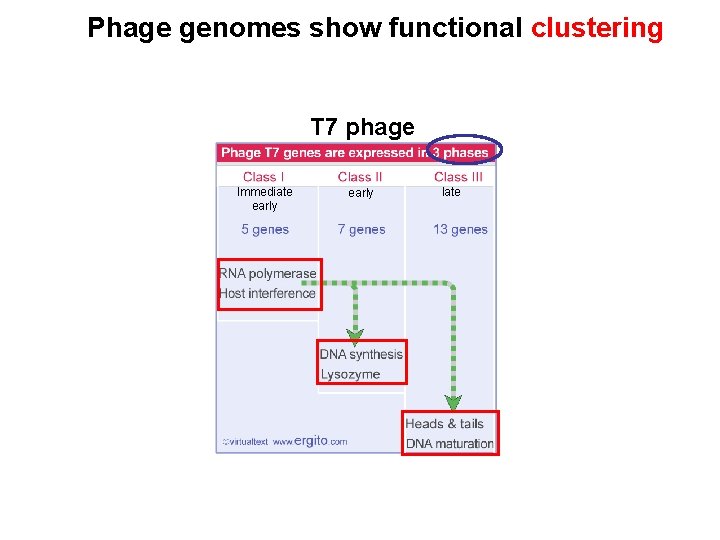
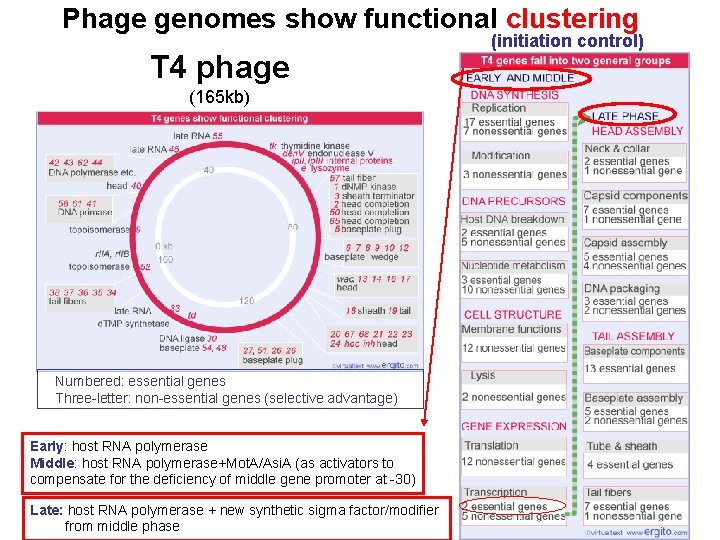
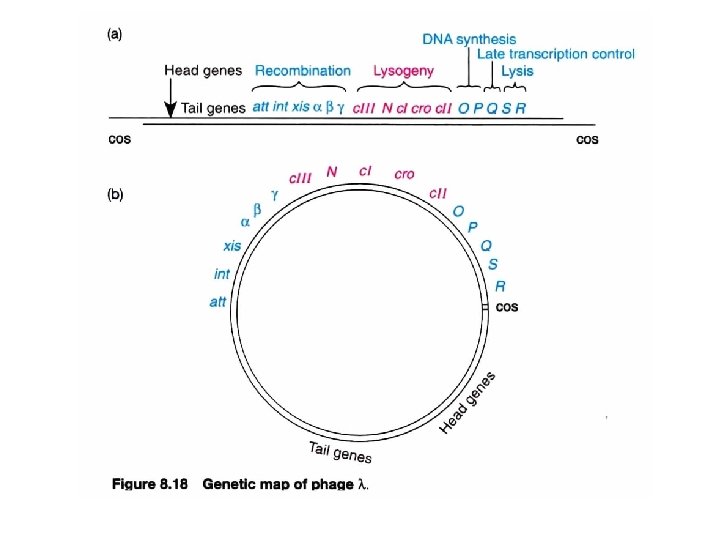
Phage genomes show functional clustering T 7 phage Immediate early late

Phage genomes show functional clustering T 4 phage (165 kb) Numbered: essential genes Three-letter: non-essential genes (selective advantage) Early: host RNA polymerase Middle: host RNA polymerase+Mot. A/Asi. A (as activators to compensate for the deficiency of middle gene promoter at -30) Late: host RNA polymerase + new synthetic sigma factor/modifier from middle phase (initiation control)


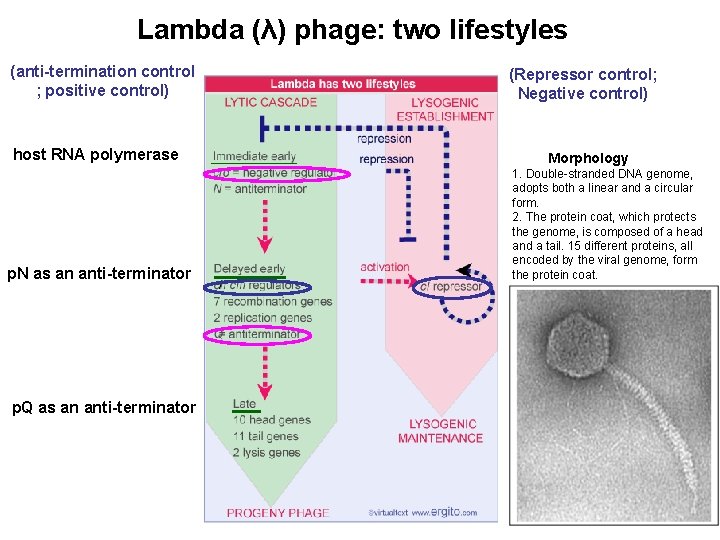
Lambda (λ) phage: two lifestyles (anti-termination control ; positive control) host RNA polymerase p. N as an anti-terminator p. Q as an anti-terminator (Repressor control; Negative control) Morphology 1. Double-stranded DNA genome, adopts both a linear and a circular form. 2. The protein coat, which protects the genome, is composed of a head and a tail. 15 different proteins, all encoded by the viral genome, form the protein coat.



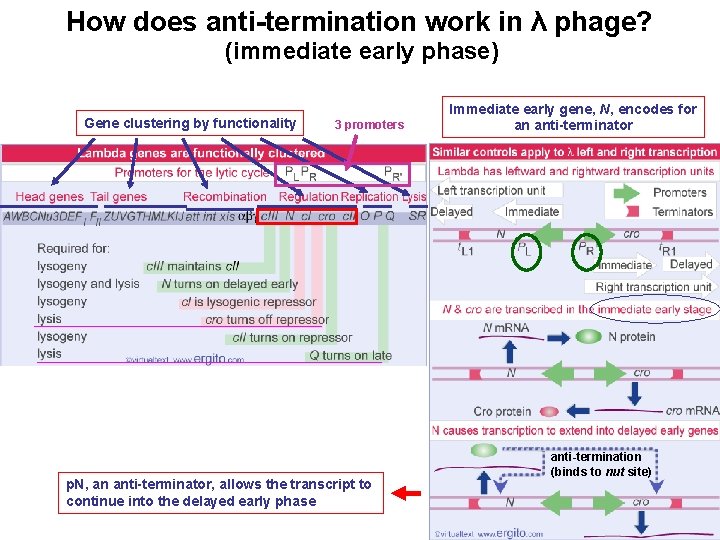
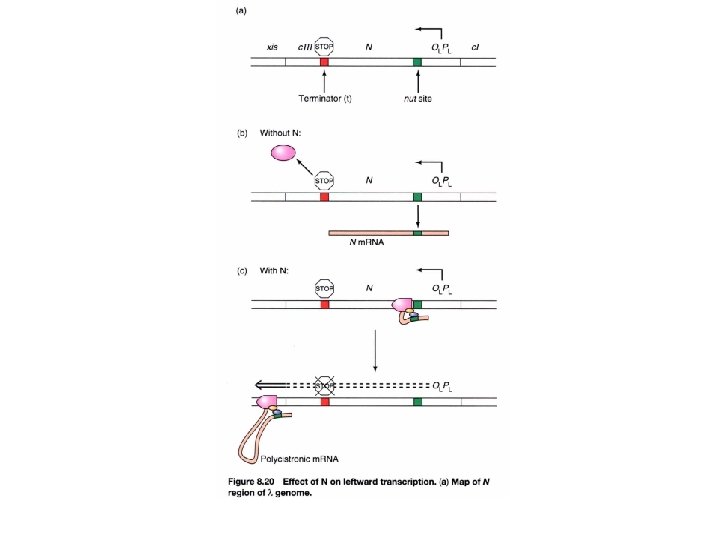
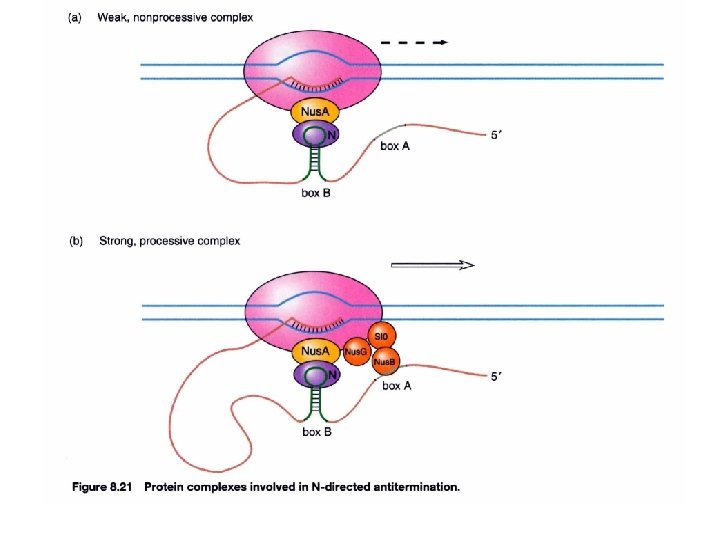
How does anti-termination work in λ phage? (immediate early phase) Gene clustering by functionality 3 promoters p. N, an anti-terminator, allows the transcript to continue into the delayed early phase Immediate early gene, N, encodes for an anti-terminator anti-termination (binds to nut site)


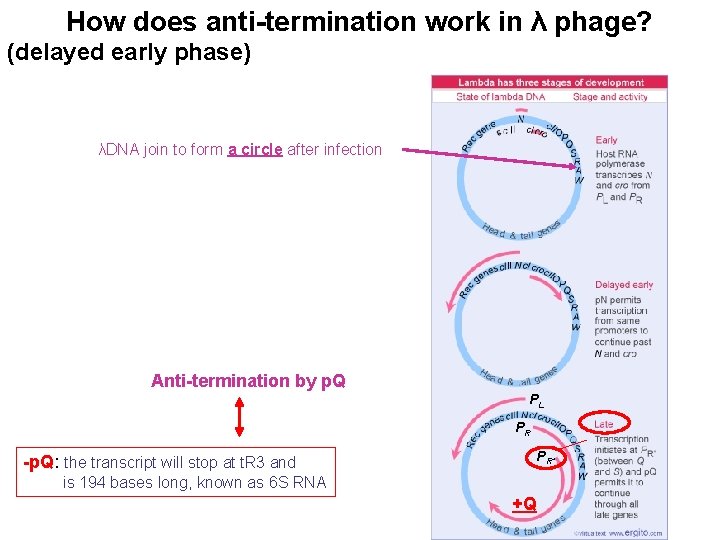
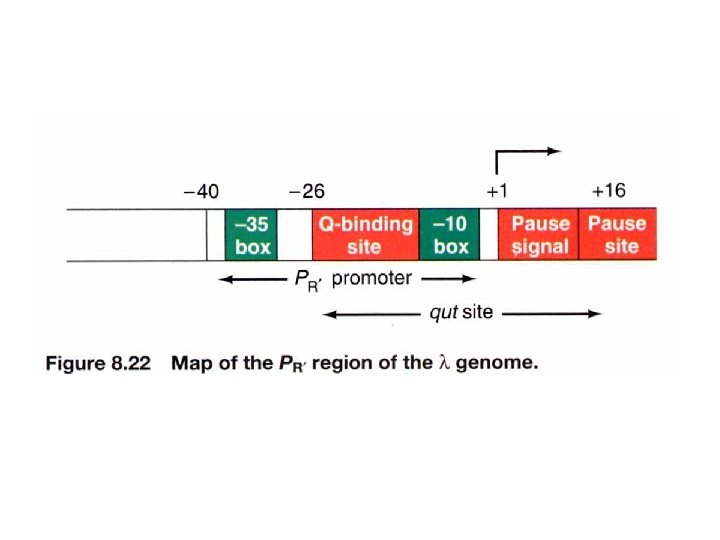
How does anti-termination work in λ phage? (delayed early phase) λDNA join to form a circle after infection Anti-termination by p. Q PL PR PR’ -p. Q: the transcript will stop at t. R 3 and is 194 bases long, known as 6 S RNA +Q




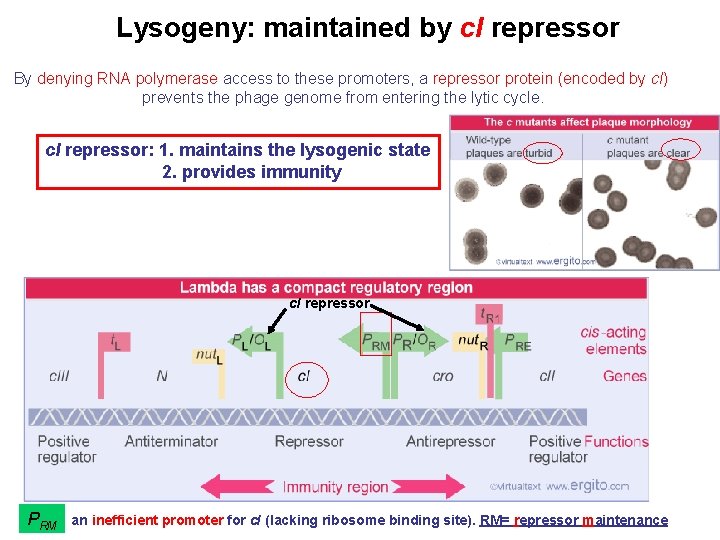
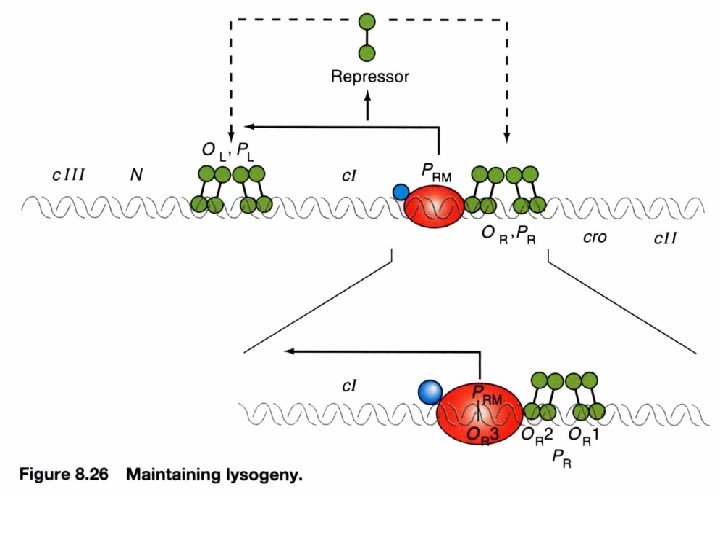
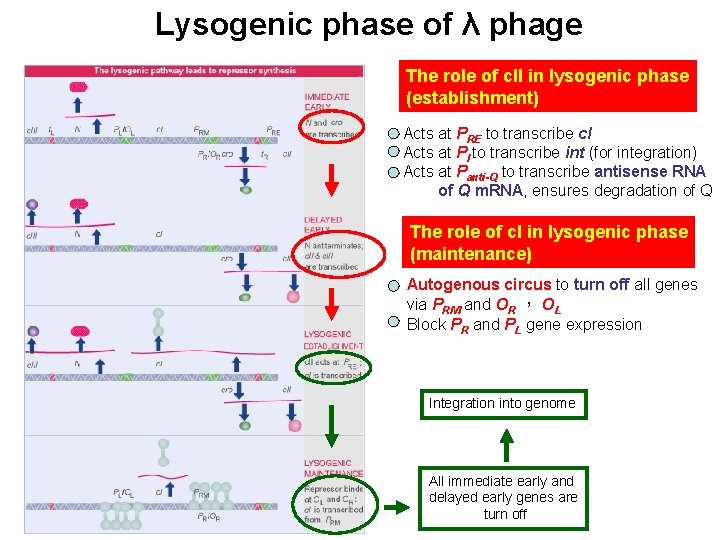
Lysogeny: maintained by c. I repressor By denying RNA polymerase access to these promoters, a repressor protein (encoded by c. I) prevents the phage genome from entering the lytic cycle. c. I repressor: 1. maintains the lysogenic state 2. provides immunity c. I repressor PRM an inefficient promoter for c. I (lacking ribosome binding site). RM= repressor maintenance

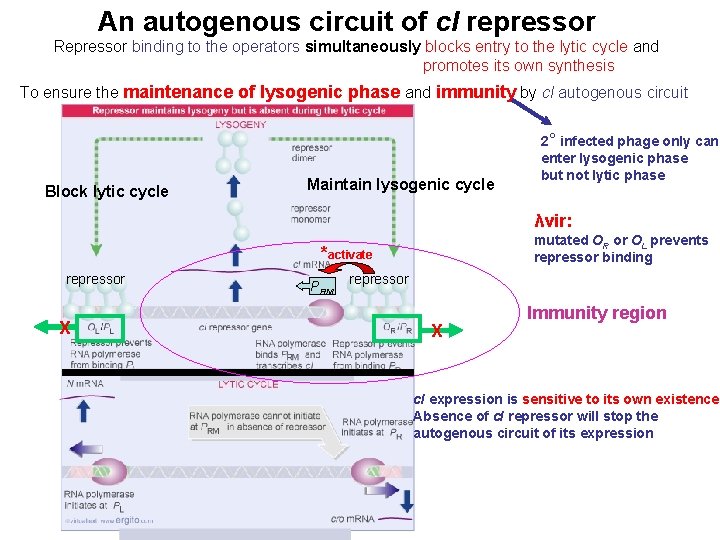
An autogenous circuit of c. I repressor Repressor binding to the operators simultaneously blocks entry to the lytic cycle and promotes its own synthesis To ensure the maintenance of lysogenic phase and immunity by c. I autogenous circuit Block lytic cycle Maintain lysogenic cycle 2° infected phage only can enter lysogenic phase but not lytic phase λvir: mutated OR or OL prevents repressor binding *activate repressor X PRM repressor X Immunity region c. I expression is sensitive to its own existence Absence of c. I repressor will stop the autogenous circuit of its expression

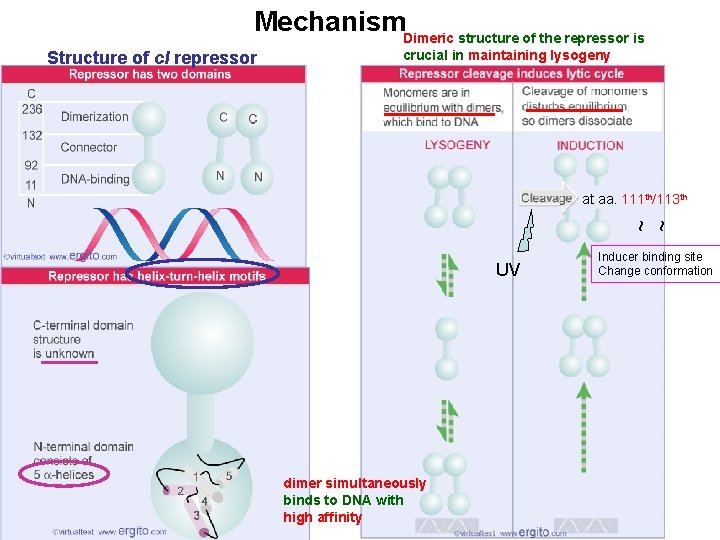
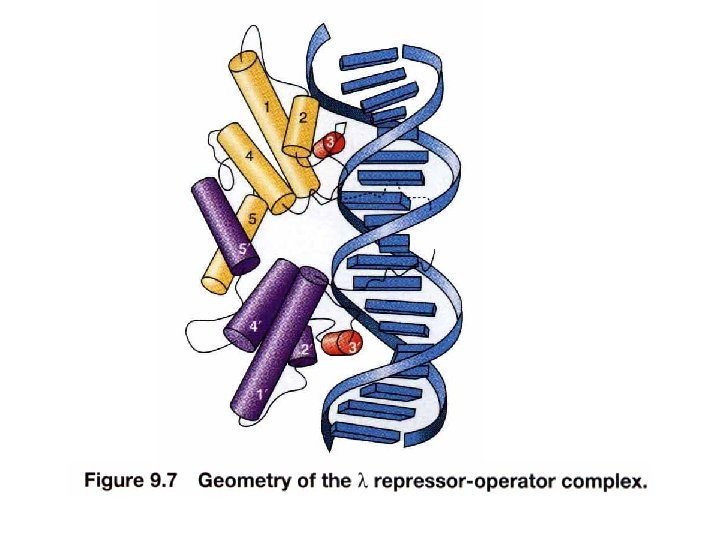
Mechanism. Dimeric structure of the repressor is crucial in maintaining lysogeny at aa. 111 th/113 th ~ ~ Structure of c. I repressor UV dimer simultaneously binds to DNA with high affinity Inducer binding site Change conformation

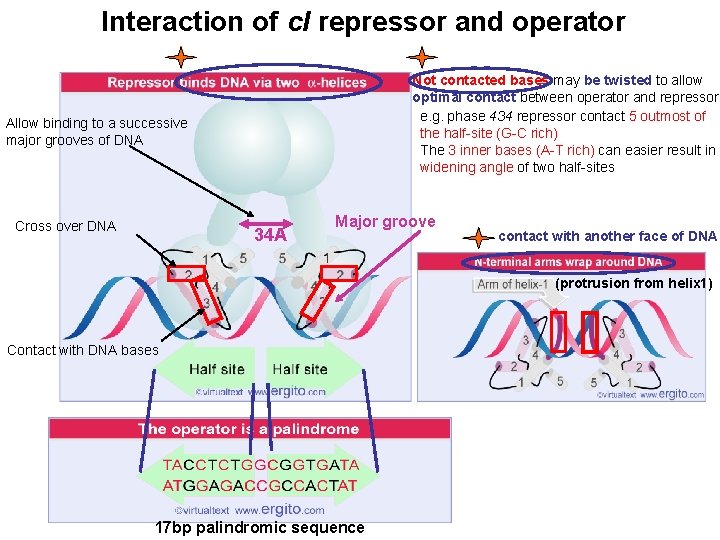
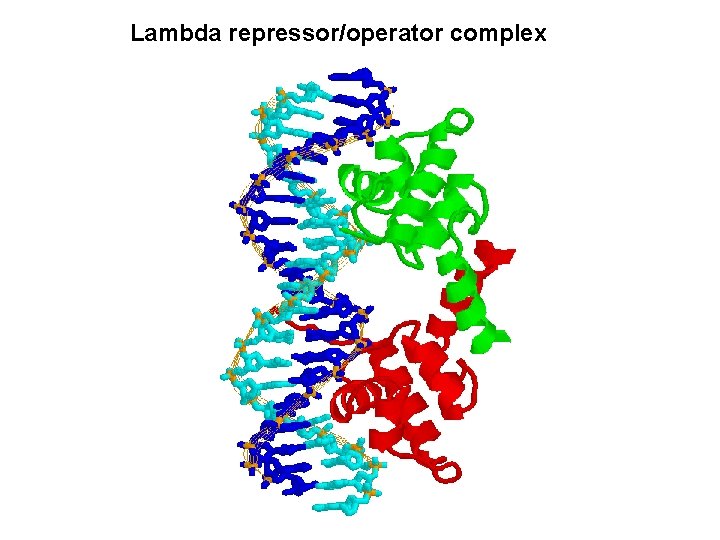
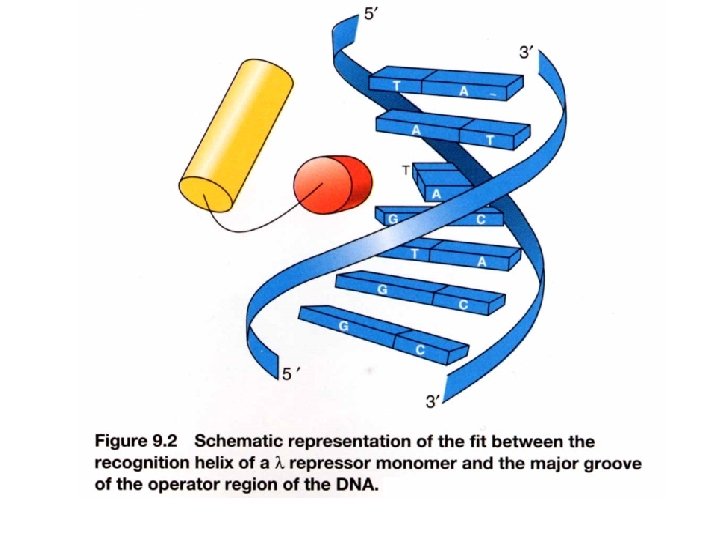
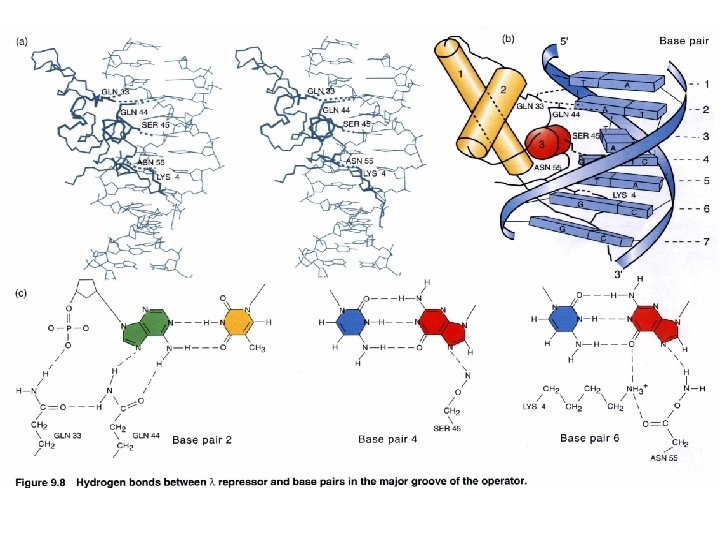
Interaction of c. I repressor and operator Not contacted bases may be twisted to allow optimal contact between operator and repressor e. g. phase 434 repressor contact 5 outmost of the half-site (G-C rich) The 3 inner bases (A-T rich) can easier result in widening angle of two half-sites Allow binding to a successive major grooves of DNA Cross over DNA 34 A Major groove contact with another face of DNA (protrusion from helix 1) Contact with DNA bases 17 bp palindromic sequence

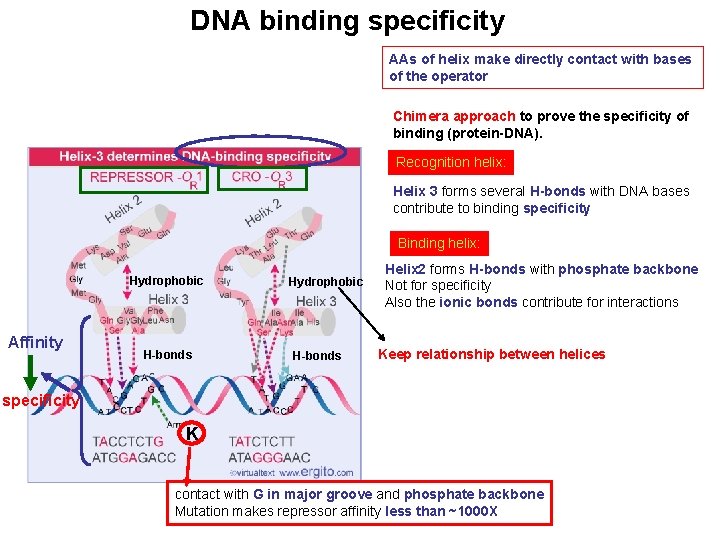
DNA binding specificity AAs of helix make directly contact with bases of the operator Chimera approach to prove the specificity of binding (protein-DNA). Recognition helix: Helix 3 forms several H-bonds with DNA bases contribute to binding specificity Binding helix: Hydrophobic Affinity H-bonds Hydrophobic H-bonds Helix 2 forms H-bonds with phosphate backbone Not for specificity Also the ionic bonds contribute for interactions Keep relationship between helices specificity K contact with G in major groove and phosphate backbone Mutation makes repressor affinity less than ~1000 X

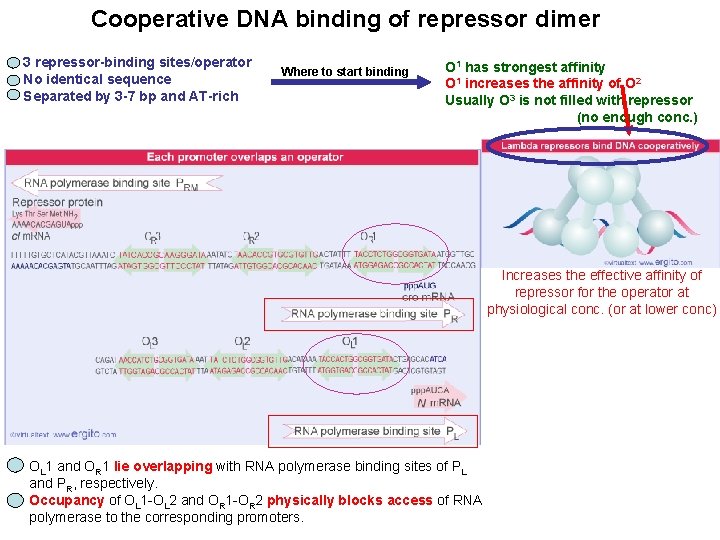
Cooperative DNA binding of repressor dimer 3 repressor-binding sites/operator No identical sequence Separated by 3 -7 bp and AT-rich Where to start binding O 1 has strongest affinity O 1 increases the affinity of O 2 Usually O 3 is not filled with repressor (no enough conc. ) Increases the effective affinity of repressor for the operator at physiological conc. (or at lower conc) OL 1 and OR 1 lie overlapping with RNA polymerase binding sites of PL and PR, respectively. Occupancy of OL 1 -OL 2 and OR 1 -OR 2 physically blocks access of RNA polymerase to the corresponding promoters.

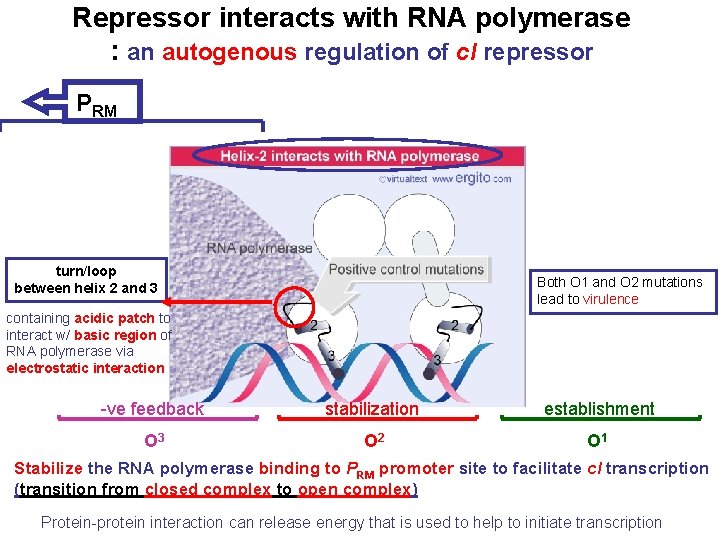
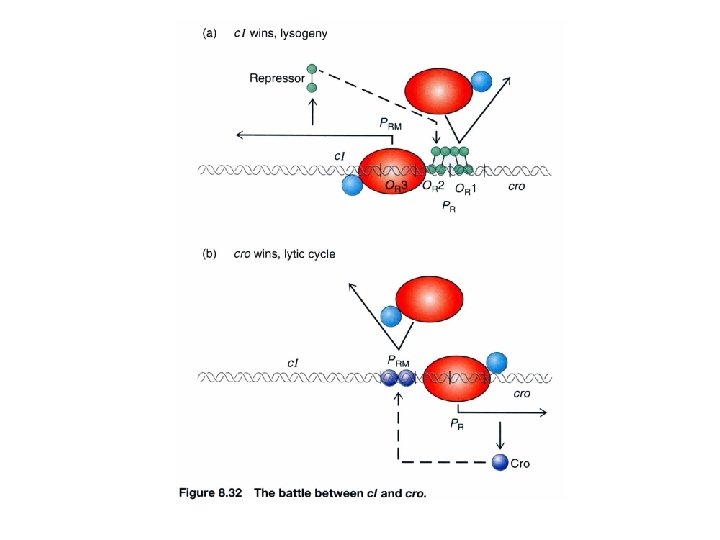
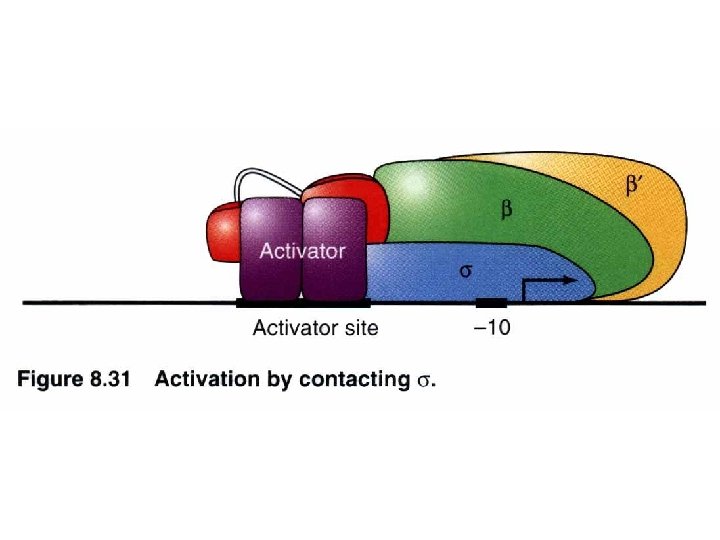
Repressor interacts with RNA polymerase : an autogenous regulation of c. I repressor PRM turn/loop between helix 2 and 3 Both O 1 and O 2 mutations lead to virulence containing acidic patch to interact w/ basic region of RNA polymerase via electrostatic interaction -ve feedback stabilization establishment O 3 O 2 O 1 Stabilize the RNA polymerase binding to PRM promoter site to facilitate c. I transcription (transition from closed complex to open complex) Protein-protein interaction can release energy that is used to help to initiate transcription

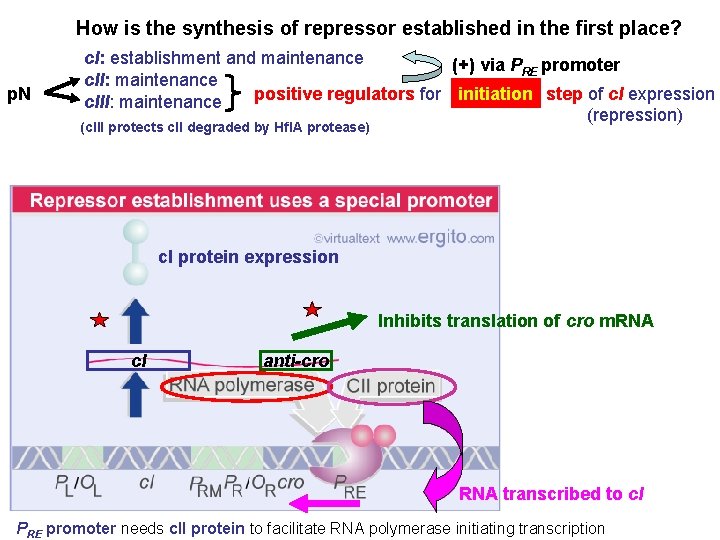
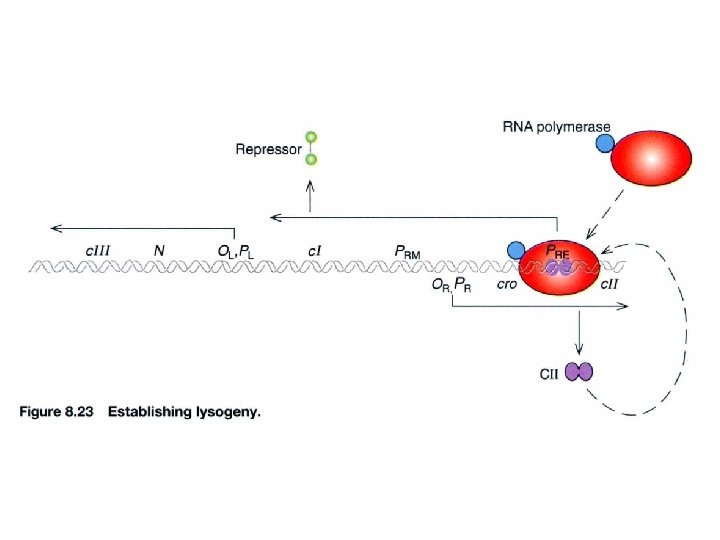
How is the synthesis of repressor established in the first place? p. N c. I: establishment and maintenance (+) via PRE promoter c. II: maintenance initiation step of c. I expression positive regulators for initiation c. III: maintenance (repression) (c. III protects c. II degraded by Hfl. A protease) cl protein expression Inhibits translation of cro m. RNA c. I anti-cro RNA transcribed to c. I PRE promoter needs c. II protein to facilitate RNA polymerase initiating transcription

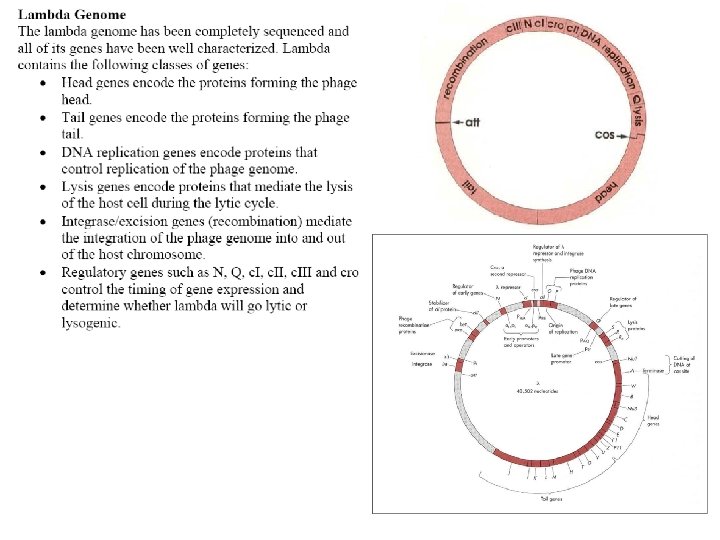
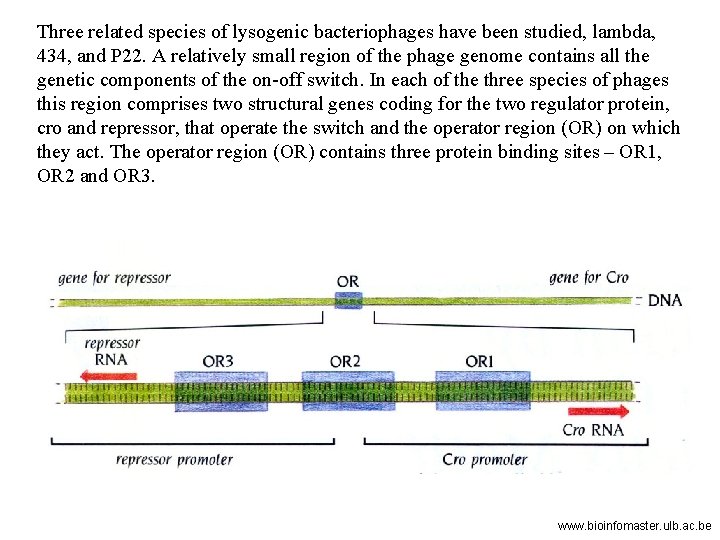
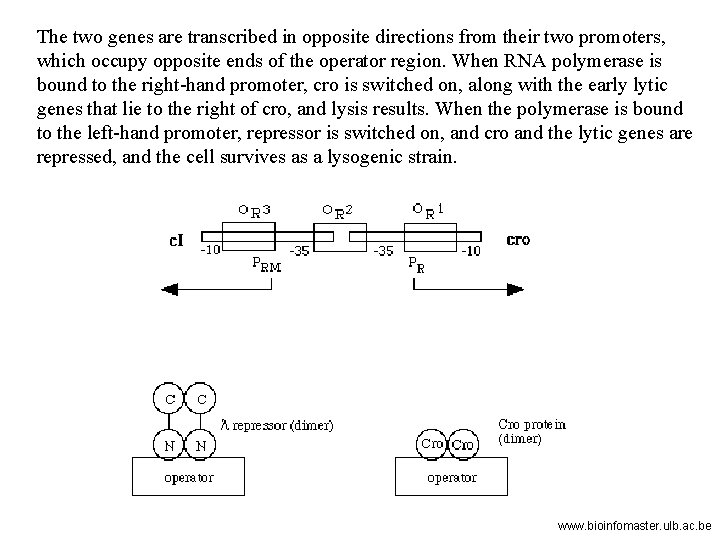
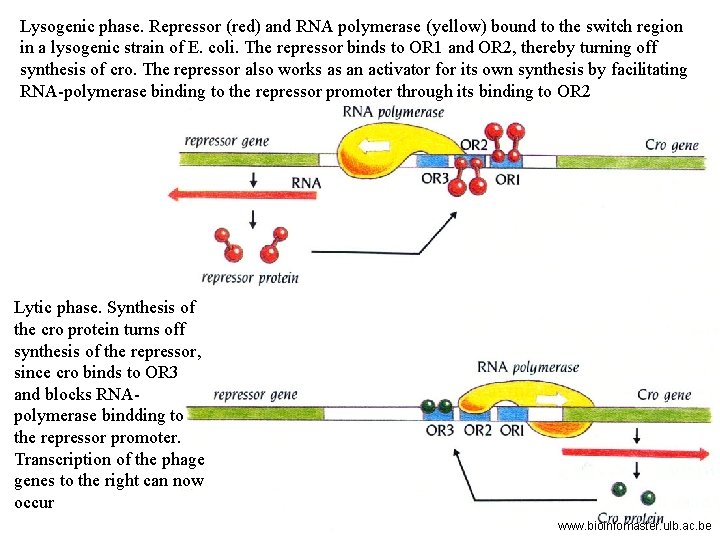
Three related species of lysogenic bacteriophages have been studied, lambda, 434, and P 22. A relatively small region of the phage genome contains all the genetic components of the on-off switch. In each of the three species of phages this region comprises two structural genes coding for the two regulator protein, cro and repressor, that operate the switch and the operator region (OR) on which they act. The operator region (OR) contains three protein binding sites – OR 1, OR 2 and OR 3. www. bioinfomaster. ulb. ac. be

The two genes are transcribed in opposite directions from their two promoters, which occupy opposite ends of the operator region. When RNA polymerase is bound to the right-hand promoter, cro is switched on, along with the early lytic genes that lie to the right of cro, and lysis results. When the polymerase is bound to the left-hand promoter, repressor is switched on, and cro and the lytic genes are repressed, and the cell survives as a lysogenic strain. www. bioinfomaster. ulb. ac. be

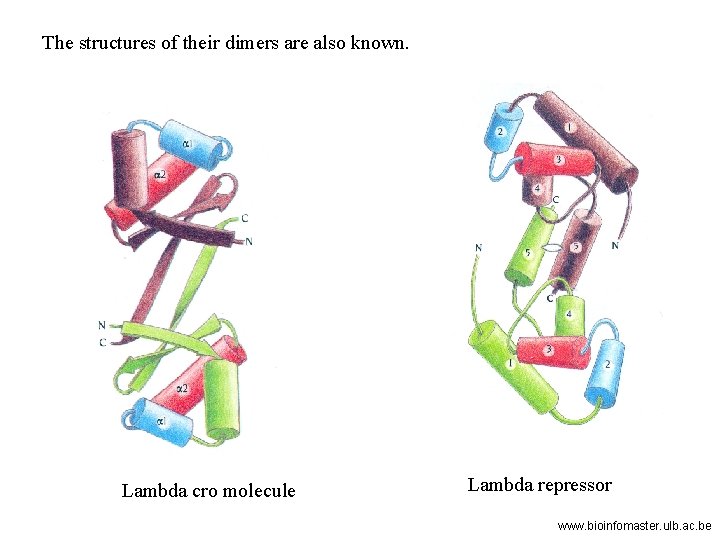
The structures of their dimers are also known. Lambda cro molecule Lambda repressor www. bioinfomaster. ulb. ac. be

Lambda repressor/operator complex

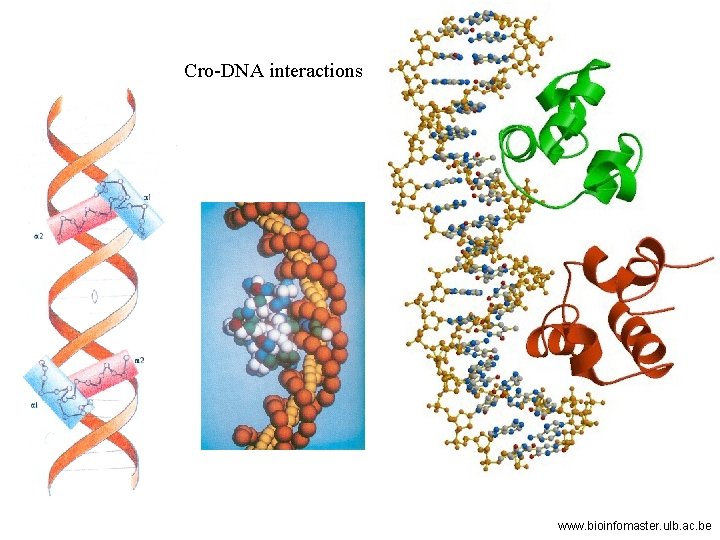
Cro-DNA interactions www. bioinfomaster. ulb. ac. be

Lysogenic phase. Repressor (red) and RNA polymerase (yellow) bound to the switch region in a lysogenic strain of E. coli. The repressor binds to OR 1 and OR 2, thereby turning off synthesis of cro. The repressor also works as an activator for its own synthesis by facilitating RNA-polymerase binding to the repressor promoter through its binding to OR 2 Lytic phase. Synthesis of the cro protein turns off synthesis of the repressor, since cro binds to OR 3 and blocks RNApolymerase bindding to the repressor promoter. Transcription of the phage genes to the right can now occur www. bioinfomaster. ulb. ac. be



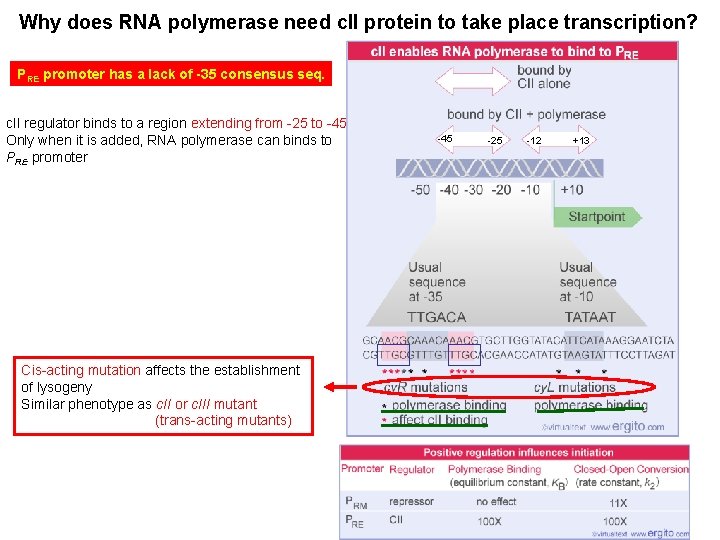
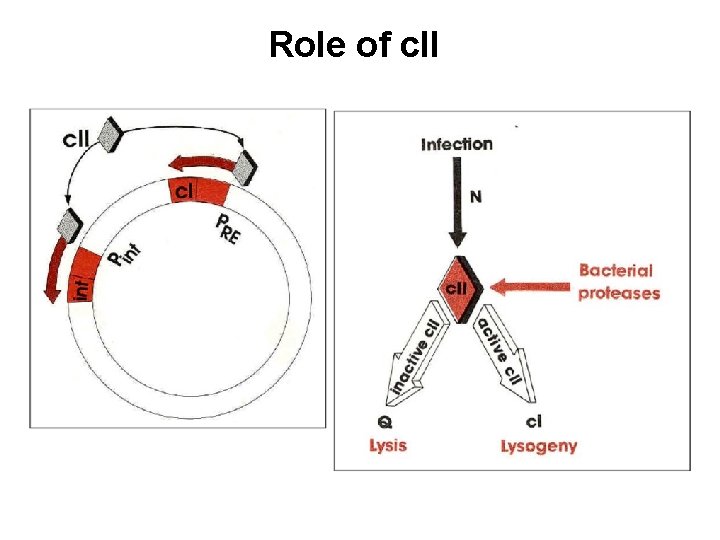
Why does RNA polymerase need c. II protein to take place transcription? PRE promoter has a lack of -35 consensus seq. c. II regulator binds to a region extending from -25 to -45 Only when it is added, RNA polymerase can binds to PRE promoter Cis-acting mutation affects the establishment of lysogeny Similar phenotype as c. II or c. III mutant (trans-acting mutants) -45 -25 -12 +13

Lysogenic phase of λ phage The role of c. II in lysogenic phase (establishment) Acts at PRE to transcribe c. I Acts at PI to transcribe int (for integration) Acts at Panti-Q to transcribe antisense RNA of Q m. RNA, ensures degradation of Q The role of c. I in lysogenic phase (maintenance) Autogenous circus to turn off all genes via PRM and OR , OL Block PR and PL gene expression Integration into genome All immediate early and delayed early genes are turn off

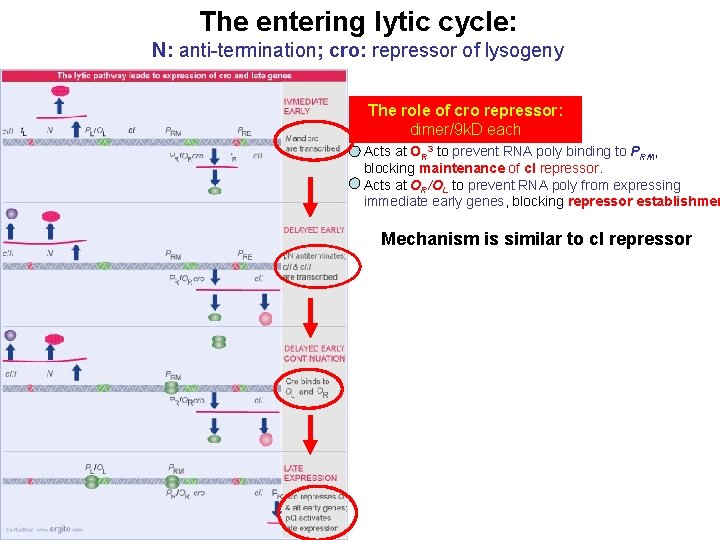
The entering lytic cycle: N: anti-termination; cro: repressor of lysogeny The role of cro repressor: dimer/9 k. D each Acts at OR 3 to prevent RNA poly binding to PRM, blocking maintenance of c. I repressor. Acts at OR/OL to prevent RNA poly from expressing immediate early genes, blocking repressor establishmen Mechanism is similar to c. I repressor

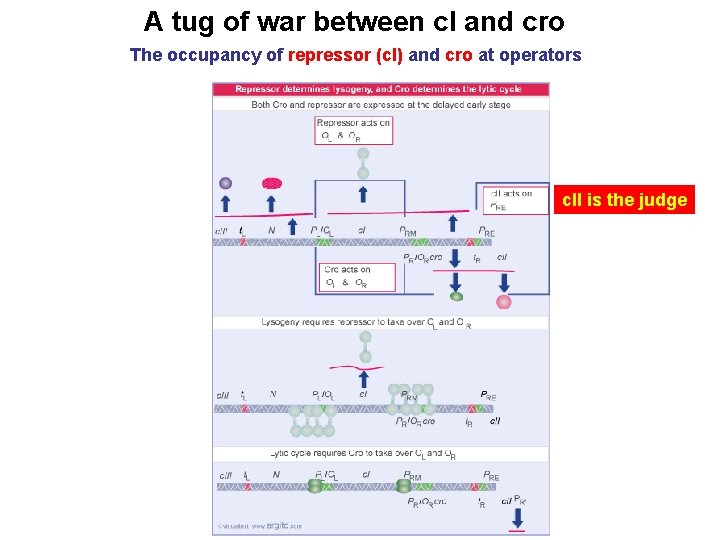
A tug of war between c. I and cro The occupancy of repressor (c. I) and cro at operators c. II is the judge



Role of c. II

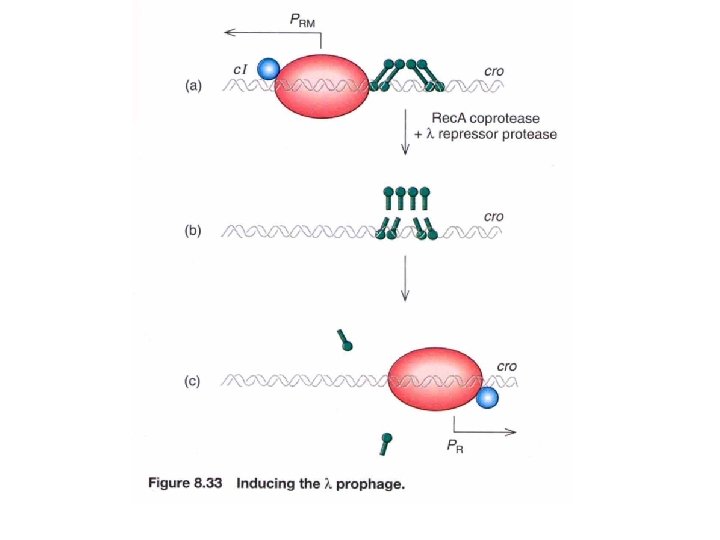
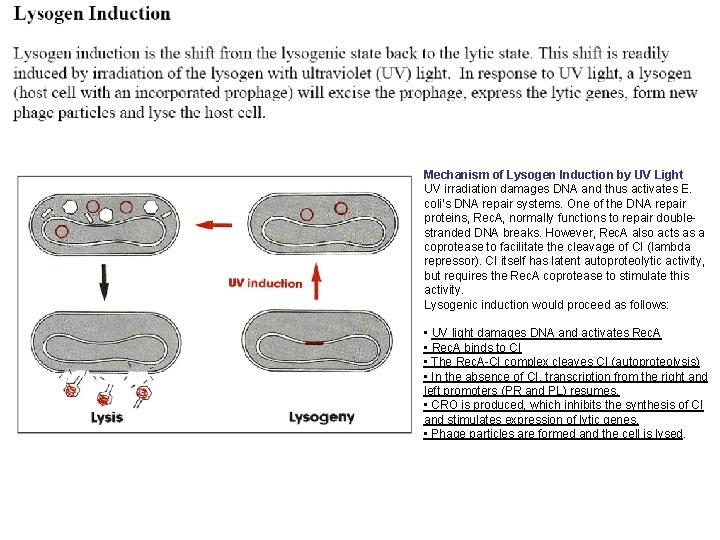
Mechanism of Lysogen Induction by UV Light UV irradiation damages DNA and thus activates E. coli’s DNA repair systems. One of the DNA repair proteins, Rec. A, normally functions to repair doublestranded DNA breaks. However, Rec. A also acts as a coprotease to facilitate the cleavage of CI (lambda repressor). CI itself has latent autoproteolytic activity, but requires the Rec. A coprotease to stimulate this activity. Lysogenic induction would proceed as follows: • UV light damages DNA and activates Rec. A • Rec. A binds to CI • The Rec. A-CI complex cleaves CI (autoproteolysis) • In the absence of CI, transcription from the right and left promoters (PR and PL) resumes. • CRO is produced, which inhibits the synthesis of CI and stimulates expression of lytic genes. • Phage particles are formed and the cell is lysed.


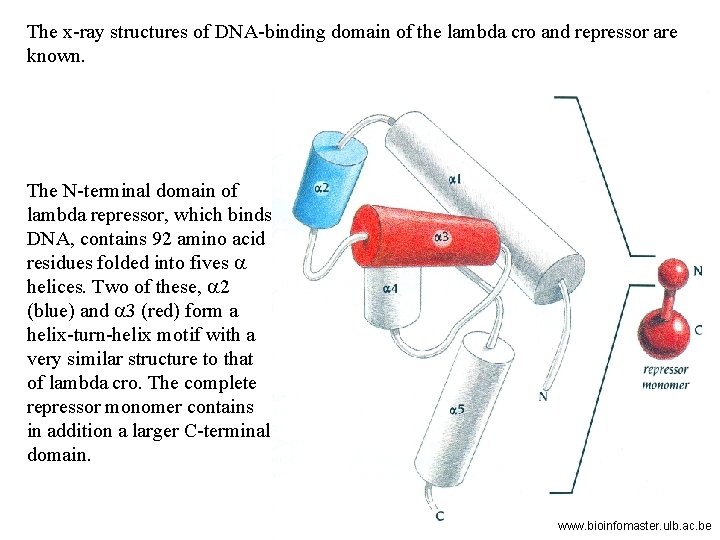
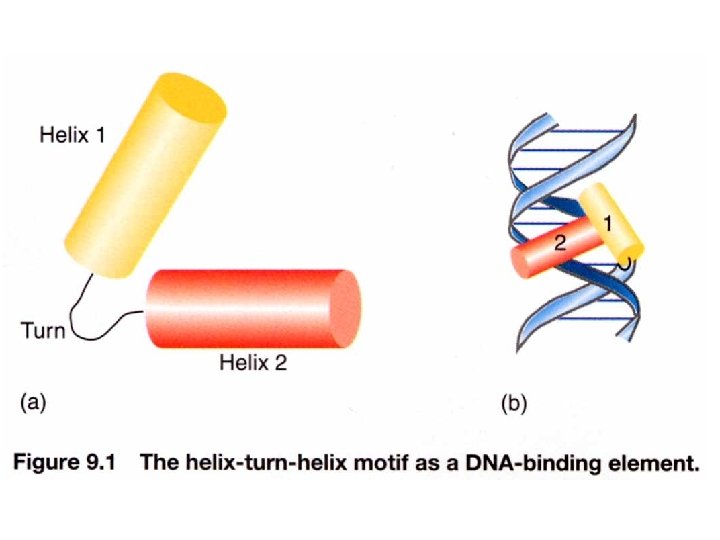
The x-ray structures of DNA-binding domain of the lambda cro and repressor are known. The N-terminal domain of lambda repressor, which binds DNA, contains 92 amino acid residues folded into fives a helices. Two of these, a 2 (blue) and a 3 (red) form a helix-turn-helix motif with a very similar structure to that of lambda cro. The complete repressor monomer contains in addition a larger C-terminal domain. www. bioinfomaster. ulb. ac. be

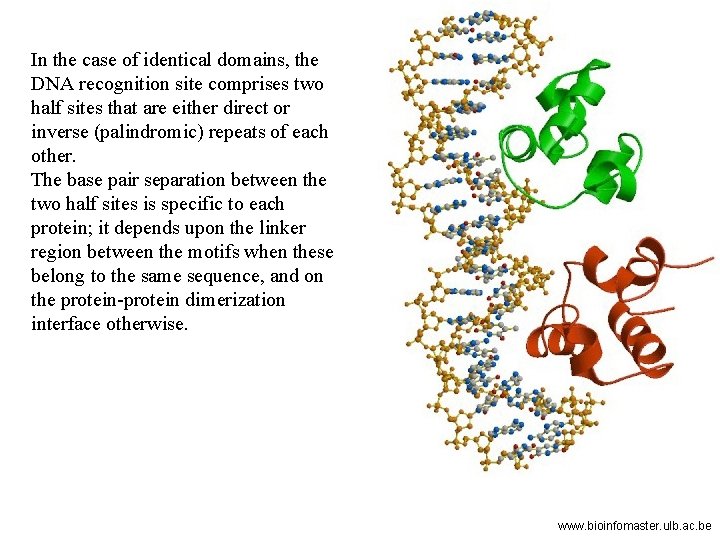
In the case of identical domains, the DNA recognition site comprises two half sites that are either direct or inverse (palindromic) repeats of each other. The base pair separation between the two half sites is specific to each protein; it depends upon the linker region between the motifs when these belong to the same sequence, and on the protein-protein dimerization interface otherwise. www. bioinfomaster. ulb. ac. be

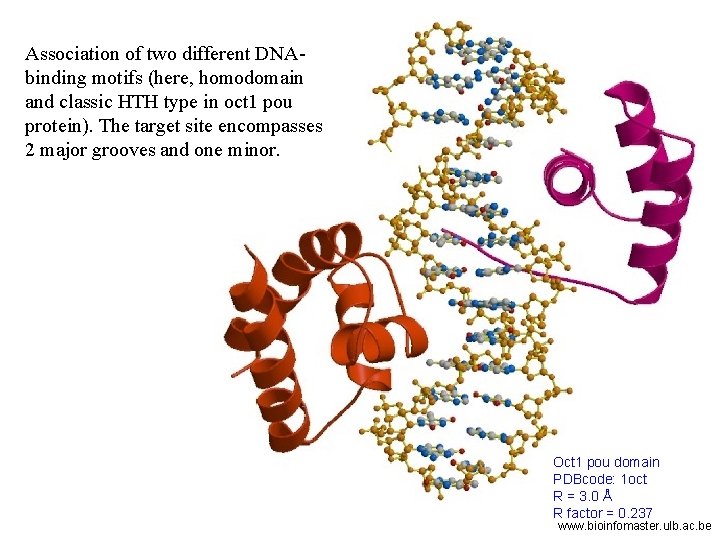
Association of two different DNAbinding motifs (here, homodomain and classic HTH type in oct 1 pou protein). The target site encompasses 2 major grooves and one minor. Oct 1 pou domain PDBcode: 1 oct R = 3. 0 Å R factor = 0. 237 www. bioinfomaster. ulb. ac. be

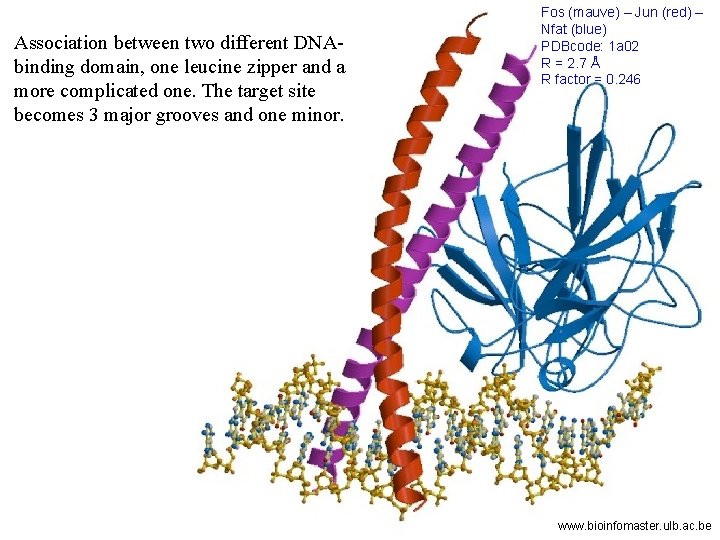
Association between two different DNAbinding domain, one leucine zipper and a more complicated one. The target site becomes 3 major grooves and one minor. Fos (mauve) – Jun (red) – Nfat (blue) PDBcode: 1 a 02 R = 2. 7 Å R factor = 0. 246 www. bioinfomaster. ulb. ac. be




